
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
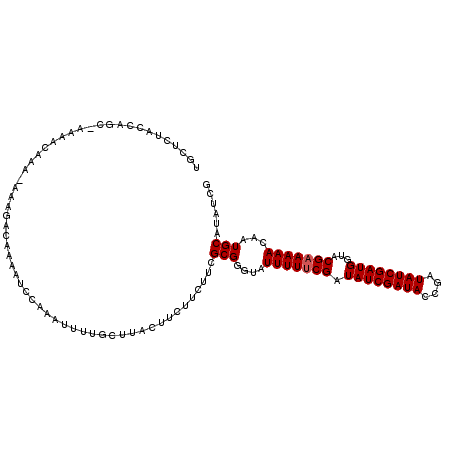
| Location | 6,916,306 – 6,916,397 |
| Length | 91 |
| Max. P | 0.882765 |

| Location | 6,916,306 – 6,916,397 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -14.57 |
| Energy contribution | -14.90 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6916306 91 + 23771897 UGCUCUACCAGA-----------------------AAAGUUUAUUUACUUCUUCUAUGCGUGUAUUUUCCGAUAUCGAUACCGAUAUCGAUGGUACGAAAAAGAAUGCAUAUCG .........(((-----------------------(((((......)))).))))((((((...(((((((((((((....)))))))).......)))))...)))))).... ( -20.61) >DroSec_CAF1 169643 113 + 1 UGCUCUAAAAGCCAAAACAAA-AAAGACAAAAUCCAAAUUUUGCUUACUUCUUCUUCGCGGGUAUUUUUCGAUAUCGAUACCGAUAUCGAUGGUACGAAAAACAAUGCAUAUCG (((.......((.........-.(((.((((((....)))))))))...........))..(((((..(((((((((....))))))))).)))))..........)))..... ( -21.90) >DroSim_CAF1 171209 114 + 1 UGCUCUACCAGCAAAAACAAAAAAAGACAAAAUCCAAAUUUUGCUUACUUUUUCUUCGCGGGUAUUUUUCGAUAUCGAUACCGAUAUCGAUGGUUCGAAAAACAAUGCAUAUCG (((..((((.((...........(((.((((((....)))))))))...........)).))))((((((((((((((((....))))))))..))))))))....)))..... ( -24.75) >consensus UGCUCUACCAGC_AAAACAAA_AAAGACAAAAUCCAAAUUUUGCUUACUUCUUCUUCGCGGGUAUUUUUCGAUAUCGAUACCGAUAUCGAUGGUACGAAAAACAAUGCAUAUCG .........................................................(((....(((((((.((((((((....))))))))...)))))))...)))...... (-14.57 = -14.90 + 0.33)


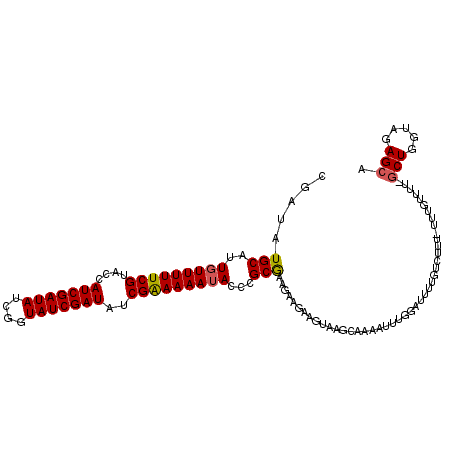
| Location | 6,916,306 – 6,916,397 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -15.01 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6916306 91 - 23771897 CGAUAUGCAUUCUUUUUCGUACCAUCGAUAUCGGUAUCGAUAUCGGAAAAUACACGCAUAGAAGAAGUAAAUAAACUUU-----------------------UCUGGUAGAGCA ...............(((.((((.(((((((((....))))))))).............((((((.((......)))))-----------------------)))))))))).. ( -19.50) >DroSec_CAF1 169643 113 - 1 CGAUAUGCAUUGUUUUUCGUACCAUCGAUAUCGGUAUCGAUAUCGAAAAAUACCCGCGAAGAAGAAGUAAGCAAAAUUUGGAUUUUGUCUUU-UUUGUUUUGGCUUUUAGAGCA .....(((.....(((((((....(((((((((....))))))))).........)))))))((((((((((((((...((((...)))).)-)))))))..))))))...))) ( -27.52) >DroSim_CAF1 171209 114 - 1 CGAUAUGCAUUGUUUUUCGAACCAUCGAUAUCGGUAUCGAUAUCGAAAAAUACCCGCGAAGAAAAAGUAAGCAAAAUUUGGAUUUUGUCUUUUUUUGUUUUUGCUGGUAGAGCA .....(((....((((((((...(((((((....))))))).))))))))((((.(((((((.((((.(((((((((....)))))).))).)))).))))))).))))..))) ( -33.60) >consensus CGAUAUGCAUUGUUUUUCGUACCAUCGAUAUCGGUAUCGAUAUCGAAAAAUACCCGCGAAGAAGAAGUAAGCAAAAUUUGGAUUUUGUCUUU_UUUGUUUU_GCUGGUAGAGCA .....(((..(((((((((....(((((((....)))))))..)))))))))...)))............................................(((.....))). (-15.01 = -15.23 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:33 2006