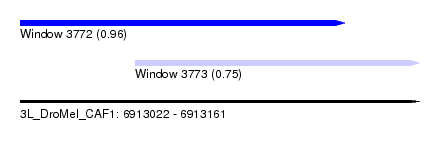
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,913,022 – 6,913,161 |
| Length | 139 |
| Max. P | 0.961146 |

| Location | 6,913,022 – 6,913,135 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.43 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6913022 113 + 23771897 UUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---CUUCUC---- ..........(((((.........)))))..((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))..---......---- ( -22.60) >DroVir_CAF1 208050 113 + 1 UUUUGCUCGCUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---UUUCUC---- ........(.(((((.........))))).)((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))..---......---- ( -23.30) >DroPse_CAF1 189863 120 + 1 UUUUGCUCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUCUCCUCU .........((((((.........)))))).((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))............... ( -23.70) >DroGri_CAF1 180063 109 + 1 UUUUGUGCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---UU-------- .........((((((.........)))))).((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))..---..-------- ( -23.70) >DroMoj_CAF1 192544 113 + 1 UUUUGCUCGCUGGCGCGUUGACAUUGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---UCUCUU---- ........(.(((((.........))))).)((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))..---......---- ( -22.00) >DroPer_CAF1 187929 120 + 1 UUUUGCUCAGUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUCUCCUCU .........((((((.........)))))).((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))............... ( -23.70) >consensus UUUUGCUCACUGGCGCGUUGACAUCGCCAUCAAAAUGCUUUGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU___CUUCUC____ ..........(((((.........)))))..((((((...(((((((((((.....(((((.(((............))).)))))..)))))))))))))))))............... (-22.57 = -22.43 + -0.14)

| Location | 6,913,062 – 6,913,161 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.79 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -15.00 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6913062 99 + 23771897 UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---CUUCUC------UC--------UGGGACGAGCUCCUCAGCUUUC-GG--- (((((((((((.....(((((.(((............))).)))))..)))))))))))........---......------.(--------(((((.....))).))).....-..--- ( -19.50) >DroVir_CAF1 208090 86 + 1 UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---UUUCUC------C---------------------UUCAGCUUUC-UG--- (((((((((((.....(((((.(((............))).)))))..)))))))))))........---......------.---------------------..........-..--- ( -15.00) >DroGri_CAF1 180103 83 + 1 UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---UU----------AU--------------------GCCAGCUUUC-UG--- (((((((((((.....(((((.(((............))).)))))..)))))))))))........---..----------..--------------------..........-..--- ( -15.00) >DroMoj_CAF1 192584 88 + 1 UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---UCUCUU------CU--------------------UUCAGCUUUCCAG--- (((((((((((.....(((((.(((............))).)))))..)))))))))))........---......------..--------------------.............--- ( -15.00) >DroAna_CAF1 189118 107 + 1 UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU---CUUCUC------CCAGCUCAGCUCGACCGAGGUCCUCUGCUUUC-GG--- (((((((((((.....(((((.(((............))).)))))..)))))))))))........---.....(------(.((..(((...(((....)))..)))..)).-))--- ( -20.10) >DroPer_CAF1 187969 107 + 1 UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUUUCUCUUCUCCUCUCAUC--------UGGG----GCUCCUCCUCUUCC-UUCCU (((((((((((.....(((((.(((............))).)))))..)))))))))))................((((.....--------.)))----).............-..... ( -19.00) >consensus UGACGUGGUUUUUGAUUGACAGGAAACUCAUAAAUAAUUCAUGUCAUUAAAUCAUGUUACAUUUUUU___CUUCUC______CC____________________CUCAGCUUUC_UG___ (((((((((((.....(((((.(((............))).)))))..)))))))))))............................................................. (-15.00 = -15.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:30 2006