| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,900,631 – 6,900,735 |
| Length | 104 |
| Max. P | 0.616796 |

| Location | 6,900,631 – 6,900,735 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.18 |
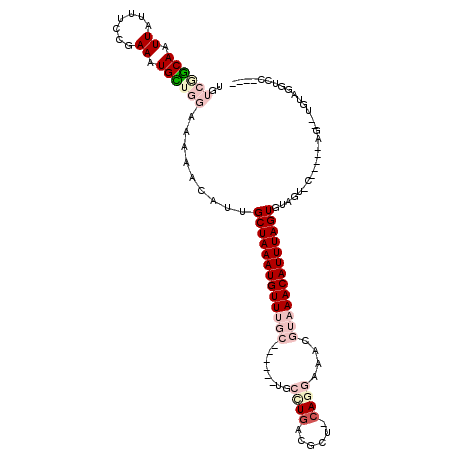
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -13.28 |
| Energy contribution | -15.58 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
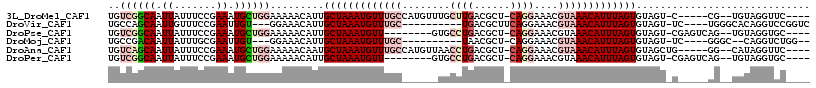
>3L_DroMel_CAF1 6900631 104 - 23771897 UGUCGGCAAUUAUUUCCGAAAUGCUGGAAAAACAUUGCUAAAUGUUUGCCAUGUUUGCUUGACGCU-CAGGAAACGUAAACAUUUAGUGUAGU-C-----CG--UGUAGGUUC---- ..((((.((....))))))..(((((((...((...(((((((((((((...((((.((((.....-)))))))))))))))))))))))..)-)-----))--.))).....---- ( -28.10) >DroVir_CAF1 192359 99 - 1 UGCCAGCAAUUGUUUCCGAAUUGU---GGAAACAUUGCUAAAUGUUUGC----------UGACGCUUCAGGAAACGUAAACAUUUAGUGUAGU-UC----UGGGCACAGGUCCGGUC ((((.(((((.(((((((.....)---)))))))))))(((((((((((----------(((....)))(....))))))))))))).)))..-.(----(((((....)))))).. ( -33.20) >DroPse_CAF1 176296 101 - 1 UGUCGGCAAUUAUUUCCGAAAUGCUGGAAAAACAUUGCUAAAUGUU--------GUGCCUGACGCU-CAGGAAACGUAAACAUUUAGUGUAGU-CGAGUCAG--UGUAGGUGC---- ..(((((.....((((((......))))))......((((((((((--------...((((.....-)))).......))))))))))...))-))).....--.........---- ( -26.60) >DroMoj_CAF1 174366 94 - 1 UGCCGACAAUUAUUUGCGAAUUGU---GGAAACAUUGCUAAAUGUUUGC----------UAACGCU-CAGGAAACGUAAACAUUUAGUGUAGU-UC----GGGC--CAGGUCUGG-- .(((..(((....)))((((((((---(....))..(((((((((((((----------....)).-..(....)..))))))))))).))))-))----))))--.........-- ( -27.30) >DroAna_CAF1 174854 105 - 1 UGUCAGCAAUUAUUUCCGAAAUGCUGGAAAAACAAUGCUAAAUGUUUGCCAUGUUAACCUGACGCU-CAGGAAACGUAAACAUUUAGUGUAGCUG-----GG--CAUAGGUUC---- (((((((.....((((((......))))))....(((((((((((((((...(((..((((.....-)))).)))))))))))))))))).))).-----))--)).......---- ( -32.60) >DroPer_CAF1 174233 101 - 1 UGUCGGCAAUUAUUUCCGAAAUGCUGGAAAAACAUUGCUAAAUGUU--------GUGCCUGACGCU-CAGGAAACGUAAACAUUUAGUGUAGU-CGAGUCAG--UGUAGGUGC---- ..(((((.....((((((......))))))......((((((((((--------...((((.....-)))).......))))))))))...))-))).....--.........---- ( -26.60) >consensus UGUCGGCAAUUAUUUCCGAAAUGCUGGAAAAACAUUGCUAAAUGUUUGC______UGCCUGACGCU_CAGGAAACGUAAACAUUUAGUGUAGU_C_____AG__UGUAGGUCC____ ..((((((.((.......)).)))))).........(((((((((((((........((((......))))....)))))))))))))............................. (-13.28 = -15.58 + 2.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:20 2006