| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,873,363 – 6,873,514 |
| Length | 151 |
| Max. P | 0.928703 |

| Location | 6,873,363 – 6,873,474 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.34 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -28.14 |
| Energy contribution | -28.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6873363 111 - 23771897 UAUCUGUGUGUUUGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUAUUGGUAGUAACCC ((((((((((..(((((........)))))..))))))))))...((((((((((........(((((....)))))........)))))))))).....(((....))). ( -34.49) >DroSec_CAF1 122055 104 - 1 ---CUGUG----UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUAUUGUUAGAAACCC ---(((((----((..(((((....)))))..)))))))......((((((((((........(((((....)))))........))))))))))................ ( -32.49) >DroSim_CAF1 128191 107 - 1 UAUCUGUG----UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUAUUGGUAGUAACCC ((((((((----((..(((((....)))))..))))))))))...((((((((((........(((((....)))))........)))))))))).....(((....))). ( -37.69) >DroEre_CAF1 138974 105 - 1 UAUC--UG----UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUAUUGGUAGAAAUCC ((((--((----((((..((((....))))..))))))))))...((((((((((........(((((....)))))........))))))))))................ ( -34.69) >consensus UAUCUGUG____UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGCAACAGGAAAUCGGACGCAUGCGUAUUGGUAGAAACCC ............((((((..(((....)))..))))))((((...((((((((((........(((((....)))))........)))))))))).))))........... (-28.14 = -28.14 + 0.00)

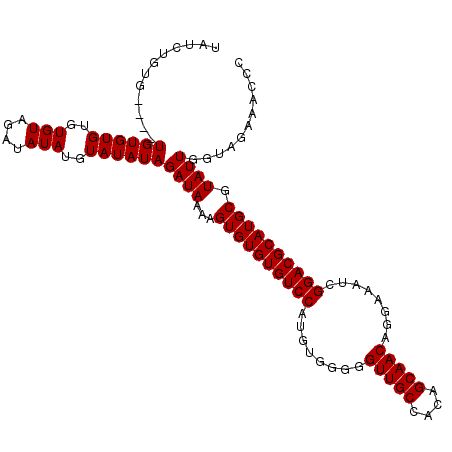

| Location | 6,873,400 – 6,873,514 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -19.08 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6873400 114 + 23771897 GCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACACAAACACACAGAUAUAUGUGUCGAGAGCACCCAACGCAAUAUUUGAAUAUUCAC (((((((......)))))................(((..((((((((((((..................))))))))))))..))).........))................. ( -20.17) >DroSim_CAF1 128228 110 + 1 GCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACACA----CACAGAUAUAUGUGUCGAGAGCACCCAACGCAAUAUUUGAAUAUUCAC (((((((......)))))................(((..((((((((((((...........----...))))))))))))..))).........))................. ( -20.44) >DroEre_CAF1 139011 108 + 1 GCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACACA----CA--GAUAUAUGUGUCGAGAGCAGGCAACGCAAUAUUUGAAUAUUCAC ..(((((......)))))((((............(((..((((((((((((...........----.)--)))))))))))..)))((.(....)))............)))). ( -22.70) >consensus GCUGUGGCAACCCCCACAUGGACACACACUUUUAUCUAUAUACAUAUAUCUACACACACACA____CACAGAUAUAUGUGUCGAGAGCACCCAACGCAAUAUUUGAAUAUUCAC (((((((......)))))................(((..(((((((((((....................)))))))))))..))).........))................. (-19.08 = -19.08 + -0.00)

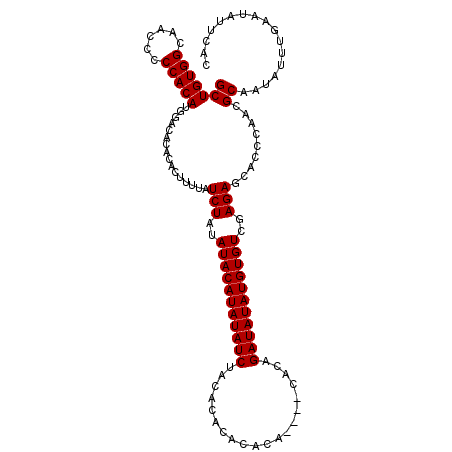

| Location | 6,873,400 – 6,873,514 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -25.83 |
| Energy contribution | -27.17 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6873400 114 - 23771897 GUGAAUAUUCAAAUAUUGCGUUGGGUGCUCUCGACACAUAUAUCUGUGUGUUUGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGC ((.(((((....)))))))(((((((((((((((((((((((((((((((..(((((........)))))..)))))))))....))))))))).....))))).)))).)))) ( -33.30) >DroSim_CAF1 128228 110 - 1 GUGAAUAUUCAAAUAUUGCGUUGGGUGCUCUCGACACAUAUAUCUGUG----UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGC ((.(((((....)))))))(((((((((((((((((((((((((((((----((..(((((....)))))..)))))))))....))))))))).....))))).)))).)))) ( -36.50) >DroEre_CAF1 139011 108 - 1 GUGAAUAUUCAAAUAUUGCGUUGCCUGCUCUCGACACAUAUAUC--UG----UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGC ((((((((....)))))(((.(.((..(....((((((((((((--((----((((..((((....))))..)))))))))....)))))))))...)..)).).))))))... ( -34.90) >consensus GUGAAUAUUCAAAUAUUGCGUUGGGUGCUCUCGACACAUAUAUCUGUG____UGUGUGUGUGUAGAUAUAUGUAUAUAGAUAAAAGUGUGUGUCCAUGUGGGGGUUGCCACAGC (.((.(((((((........))))))).)).)((((((((((((((((....((..(((((....)))))..)))))))))....)))))))))..(((((......))))).. (-25.83 = -27.17 + 1.33)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:15 2006