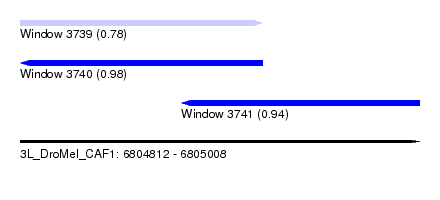
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,804,812 – 6,805,008 |
| Length | 196 |
| Max. P | 0.984234 |

| Location | 6,804,812 – 6,804,931 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -31.10 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6804812 119 + 23771897 CACUGGAGAUUUCACAGCUUUUGGCUUUGUUUGAUUUGUCAAAAUGUUAAAUGGAUCCCCGUGGAAGAUCCGUGUGUCGUAGCUGGAAAUUGGUCAGCGUGUUUGCAAGUGGCAAACGA (.((((.((((((.(((((..((((....(((((....))))).......(((((((.(....)..)))))))..)))).)))))))))))..)))).)(((((((.....))))))). ( -36.40) >DroSec_CAF1 65698 119 + 1 CACUGGAGAUUUCACAGCUUUUGGCUUUGUUUGAUUUGUCAAAAUGUUAAAUGGAUCCCCGAGGAAGAUCCGUCUGCCGUAGCUGGAAAUUGGUCAGCGUGUUUGCAAGUGGCAAACGA (.((((.((((((.(((((..((((....(((((....))))).......(((((((.(....)..)))))))..)))).)))))))))))..)))).)(((((((.....))))))). ( -38.50) >DroSim_CAF1 68308 119 + 1 CACUGGAGAUUUCACAGCUUUUGGCUUUGUUUGAUUUGCCAAAAUGUUAAAUGGAUCCCCGAGGAAGAUCCGUCUGCCGUAGCUGGAAAUUGGUCAGCGUGUUUGCAAGUGGCAAACGA (.((((.((((((.(((((((((((............))))))(((.((.(((((((.(....)..))))))).)).))))))))))))))..)))).)(((((((.....))))))). ( -37.90) >DroEre_CAF1 79192 119 + 1 CACUGGAGAUUUCACAGCUUUUGGCUUUGUUUGAUUUGUCAAAAUGUUAAAUGGAUCCCCGAGGAAGAUCCAUCUGCCGUAGCUGGAAAUUGGUCAGCGUGUUUGCAAGUGGCAAACGA (.((((.((((((.(((((..((((....(((((....))))).......(((((((.(....)..)))))))..)))).)))))))))))..)))).)(((((((.....))))))). ( -38.90) >DroYak_CAF1 70710 119 + 1 CACUGGAGAUUUCACAGCUUUUGGCUUUGUUUGAUUUGUCAAAAUGUUAAAUGGAUCCCCGAGGAAGAUCCAUCUGCCGUAGCUGGAAAUUGGUCAGCGUGUUUGCAAGUGGCAAACGA (.((((.((((((.(((((..((((....(((((....))))).......(((((((.(....)..)))))))..)))).)))))))))))..)))).)(((((((.....))))))). ( -38.90) >DroAna_CAF1 76680 104 + 1 CACUGGAGAUUUCACAGUUUUUGGCUUUGUUUGAUUUGUCGAAAUGUUGAUUUGAUUCCCCAGGAAGAUCCAGCUGGUGUAACUAGAAAUUGGUCAG---------------CAAAUGA ..((((.((..(((......(..((....(((((....)))))..))..)..))).)).))))...((.(((.((((.....))))....)))))..---------------....... ( -19.40) >consensus CACUGGAGAUUUCACAGCUUUUGGCUUUGUUUGAUUUGUCAAAAUGUUAAAUGGAUCCCCGAGGAAGAUCCAUCUGCCGUAGCUGGAAAUUGGUCAGCGUGUUUGCAAGUGGCAAACGA ..((((.((((((.(((((..((((....(((((....))))).......(((((((.(....)..)))))))..)))).)))))))))))..))))...((((((.....)))))).. (-31.10 = -31.32 + 0.21)

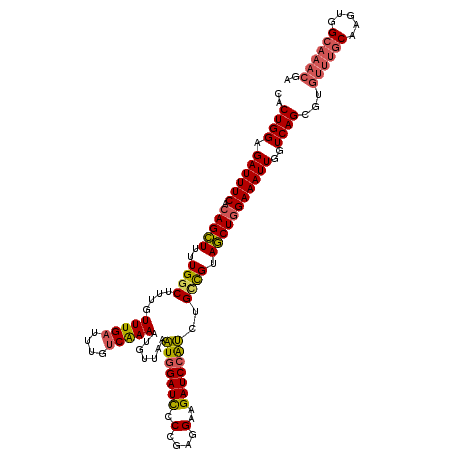
| Location | 6,804,812 – 6,804,931 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.59 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -28.06 |
| Energy contribution | -29.72 |
| Covariance contribution | 1.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6804812 119 - 23771897 UCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUACGACACACGGAUCUUCCACGGGGAUCCAUUUAACAUUUUGACAAAUCAAACAAAGCCAAAAGCUGUGAAAUCUCCAGUG ..((((((.....)))))).(((((....((((((((((.........(((((((.....))))))).........(((((....)))))..........))))).)))))...))))) ( -33.20) >DroSec_CAF1 65698 119 - 1 UCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUACGGCAGACGGAUCUUCCUCGGGGAUCCAUUUAACAUUUUGACAAAUCAAACAAAGCCAAAAGCUGUGAAAUCUCCAGUG ..((((((.....)))))).(((((....((((((((((..(((....(((((((.....))))))).........(((((....)))))....)))...))))).)))))...))))) ( -38.30) >DroSim_CAF1 68308 119 - 1 UCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUACGGCAGACGGAUCUUCCUCGGGGAUCCAUUUAACAUUUUGGCAAAUCAAACAAAGCCAAAAGCUGUGAAAUCUCCAGUG ..((((((.....)))))).(((((....((((((((((..(..(((.(((((((.....))))))).)))..)..((((((............))))))))))).)))))...))))) ( -38.80) >DroEre_CAF1 79192 119 - 1 UCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUACGGCAGAUGGAUCUUCCUCGGGGAUCCAUUUAACAUUUUGACAAAUCAAACAAAGCCAAAAGCUGUGAAAUCUCCAGUG ..((((((.....)))))).(((((....((((((((((..((((((((((((((.....))))))))))).....(((((....)))))....)))...))))).)))))...))))) ( -41.90) >DroYak_CAF1 70710 119 - 1 UCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUACGGCAGAUGGAUCUUCCUCGGGGAUCCAUUUAACAUUUUGACAAAUCAAACAAAGCCAAAAGCUGUGAAAUCUCCAGUG ..((((((.....)))))).(((((....((((((((((..((((((((((((((.....))))))))))).....(((((....)))))....)))...))))).)))))...))))) ( -41.90) >DroAna_CAF1 76680 104 - 1 UCAUUUG---------------CUGACCAAUUUCUAGUUACACCAGCUGGAUCUUCCUGGGGAAUCAAAUCAACAUUUCGACAAAUCAAACAAAGCCAAAAACUGUGAAAUCUCCAGUG .......---------------..........(((((((.....))))))).....(((((((................((....))...((.((.......)).))...))))))).. ( -14.30) >consensus UCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUACGGCAGACGGAUCUUCCUCGGGGAUCCAUUUAACAUUUUGACAAAUCAAACAAAGCCAAAAGCUGUGAAAUCUCCAGUG ..((((((.....)))))).(((((....((((((((((..(((....(((((((.....))))))).........(((((....)))))....)))...))))).)))))...))))) (-28.06 = -29.72 + 1.66)

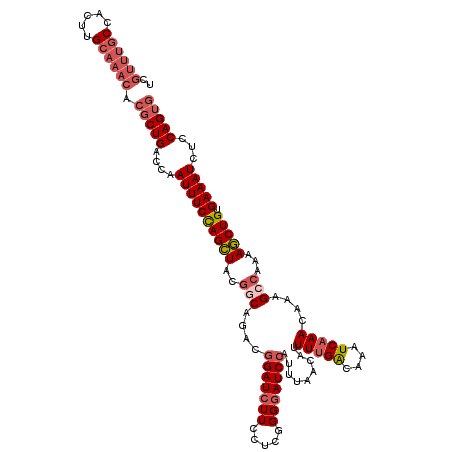
| Location | 6,804,891 – 6,805,008 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.95 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.56 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
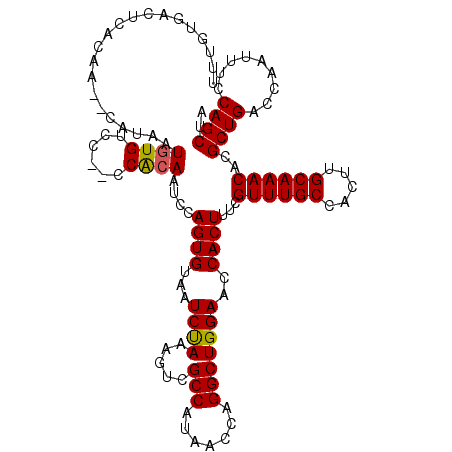
>3L_DroMel_CAF1 6804891 117 - 23771897 UUUGUCACUCACAA--CAUAAUGUGUCACCCCACAAUCCAGUGUAAUCUAGAGUCGCCAUAACCAGGCUGGAACCACUUUCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUA ...((.((((....--.....((((......))))....((......)).)))).))........((((((((........((((((.....))))))..(.....)...)))))))). ( -23.10) >DroSec_CAF1 65777 117 - 1 UUUGUGACUCACAA--CAUAAUGUGUCCCCCCACAAUCAAGUGUAAUCUACAGUCGCCAUAACCAGGCUGGAACCACUUUCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUA ...((((((((((.--.....))))......(((......)))........))))))........((((((((........((((((.....))))))..(.....)...)))))))). ( -26.20) >DroSim_CAF1 68387 115 - 1 UUUGUGACUCACAA--CAUAAUGUGUCC--CCACAAUCCAGUGUAAUCUAGAGUCGCCAUAACCAGGCUGGAACCACUUUCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUA ...(((((((....--.....((((...--.))))....((......)).)))))))........((((((((........((((((.....))))))..(.....)...)))))))). ( -28.40) >DroEre_CAF1 79271 98 - 1 CC-------------------UGUGUCC--CCACAAUCCAGUGUAAUCUAAAGUCGCCUUAAACAGGCUGGAACCACUUUCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUA ..-------------------((((...--.))))...((((((.....(((((.((((.....)))).(....)))))).((((((.....))))))))))))............... ( -21.50) >DroYak_CAF1 70789 117 - 1 UCACAAACUCACAACACAUAAUGUGUCC--CCGAAAUCGAGUGUAAUCCAAAGUCGCCAUAAACAGGCUGGAACCACUUUCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUA .............((((.....))))..--........(((((...((((.....(((.......)))))))..)))))..((((((.....))))))..((((.........)))).. ( -23.70) >consensus UUUGUGACUCACAA__CAUAAUGUGUCC__CCACAAUCCAGUGUAAUCUAAAGUCGCCAUAACCAGGCUGGAACCACUUUCGUUUGCCACUUGCAAACACGCUGACCAAUUUCCAGCUA .....................((((......))))....((((...((((.....(((.......)))))))..))))...((((((.....))))))..((((.........)))).. (-19.68 = -19.56 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:59 2006