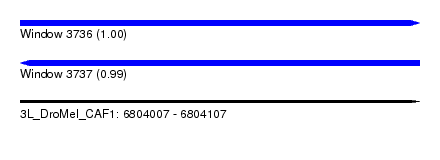
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,804,007 – 6,804,107 |
| Length | 100 |
| Max. P | 0.996172 |

| Location | 6,804,007 – 6,804,107 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.53 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
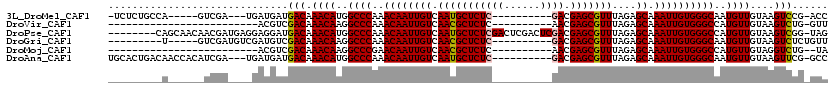
>3L_DroMel_CAF1 6804007 100 + 23771897 GGU-CGGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUC----------GAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCA---UCGAC-----UGGCAGAGA- (((-(((((....(((((.((((((((((((((((..((....)).----------.)))))).....))))))..)))).))))).))).......---.))))-----)........- ( -29.40) >DroVir_CAF1 91711 84 + 1 AAC-CAGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUU----------GAGAGCGUUGACAAUUGUUUGGGCCUUGUUUGUCGACGU------------------------- ...-..(((..((((...(((((((((((.(.((..(((((((...----------..))))))))))))))))..)))))))))..))).....------------------------- ( -30.70) >DroPse_CAF1 78962 111 + 1 CUA-CCGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUCGAGUCGAGUCGAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCCUCCUCAUCGUUGUUGCUG-------- ...-..(((....((((((((((((((((((((((....(((((......)))))..)))))).....))))))..)))))))))).)))......................-------- ( -38.20) >DroGri_CAF1 78005 96 + 1 AACAGAGACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUC----------GAGAGCGUUGACAAUUGUUUGGGCCUUGUUUGUCGACAUCGACAUCGAC-----A--------- ....((.......((((..((((((((((.(.((..(((((((...----------..))))))))))))))))..))))..))))(((((....)))))))...-----.--------- ( -30.00) >DroMoj_CAF1 84434 83 + 1 UA--CAGACCUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUU----------GAGAGCGUUGACAAUUGUUCGGGCCUUGUUUGUCGACGU------------------------- ..--..(((..((((...(((((((((((.(.((..(((((((...----------..))))))))))))))))..)))))))))..))).....------------------------- ( -29.80) >DroAna_CAF1 75817 106 + 1 GGC-CGAACUUACAACAUUGCCCACAAUUUGCUCUAAACGCUCGUC----------GAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAUCAUCA---UCGAUGUGGUUGUCAGUGCA (((-((.....(((((((.((((((((((((((((..((....)).----------.)))))).....))))))..)))).))).))))((((....---..)))))))))......... ( -30.00) >consensus AAC_CAGACUUACAACAUGGCCCACAAUUUGCUCUAAACGCUCGUC__________GAGAGCAUUGACAAUUGUUUGGGCCAUGUUUGUCAACAUC____UCGA______U_________ ......(((....((((..((((((((((((((((......................)))))).....))))))..))))..)))).))).............................. (-19.62 = -19.53 + -0.08)


| Location | 6,804,007 – 6,804,107 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -23.67 |
| Energy contribution | -24.03 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6804007 100 - 23771897 -UCUCUGCCA-----GUCGA---UGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUC----------GACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCCG-ACC -.........-----((((.---.(((....((((.(((.((((..((((((.....((((((.----------(((....))).)))))))))))))))).)))))))..)))))-)). ( -31.70) >DroVir_CAF1 91711 84 - 1 -------------------------ACGUCGACAAACAAGGCCCAAACAAUUGUCAACGCUCUC----------AACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCUG-GUU -------------------------.....(((.((((.(((((..((((((((.(((((((..----------...)))))))....)).))))))))))).))))....)))..-... ( -29.80) >DroPse_CAF1 78962 111 - 1 --------CAGCAACAACGAUGAGGAGGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUCGACUCGACUCGACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAAGUCGG-UAG --------...................(((....((((((((((..((((((((.(((((((((((......)))).)))))))....)).))))))))))))))))....)))..-... ( -37.60) >DroGri_CAF1 78005 96 - 1 ---------U-----GUCGAUGUCGAUGUCGACAAACAAGGCCCAAACAAUUGUCAACGCUCUC----------GACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCUCUGUU ---------(-----((((((......)))))))((((..((((..((((((((.(((((((..----------...)))))))....)).))))))))))..))))............. ( -32.40) >DroMoj_CAF1 84434 83 - 1 -------------------------ACGUCGACAAACAAGGCCCGAACAAUUGUCAACGCUCUC----------AACGAGCGUUUAGAGCAAAUUGUGGGCCAUGUUGUAGGUCUG--UA -------------------------.....(((.((((.(((((..((((((((.(((((((..----------...)))))))....)).))))))))))).))))....)))..--.. ( -30.90) >DroAna_CAF1 75817 106 - 1 UGCACUGACAACCACAUCGA---UGAUGAUGACAAACAUGGCCCAAACAAUUGUCAAUGCUCUC----------GACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUUCG-GCC .((.(.(((.....((((..---....))))((((.(((.((((..((((((.....((((((.----------(((....))).)))))))))))))))).)))))))..))).)-)). ( -31.80) >consensus _________A______UCGA____GAUGACGACAAACAAGGCCCAAACAAUUGUCAACGCUCUC__________GACGAGCGUUUAGAGCAAAUUGUGGGCAAUGUUGUAAGUCUG_GUC ..............................(((.((((..((((..((((((((.(((((((((((......)))).)))))))....)).))))))))))..))))....)))...... (-23.67 = -24.03 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:55 2006