
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,794,049 – 6,794,144 |
| Length | 95 |
| Max. P | 0.764637 |

| Location | 6,794,049 – 6,794,144 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -35.13 |
| Consensus MFE | -22.79 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
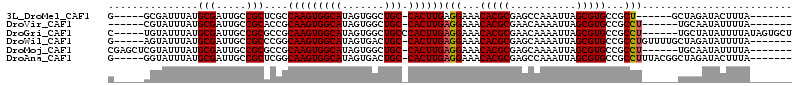
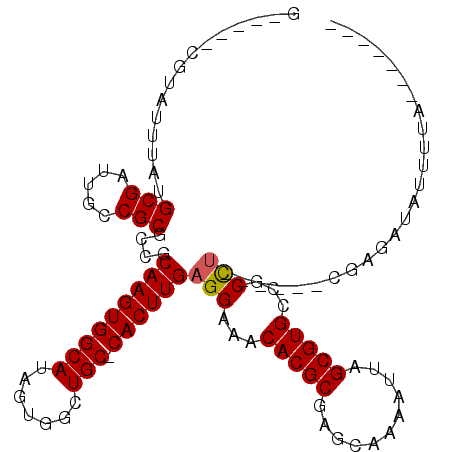
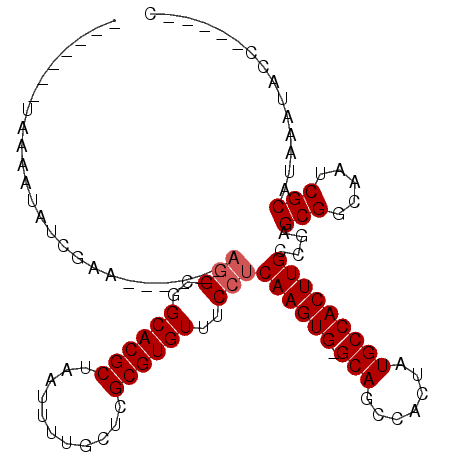
>3L_DroMel_CAF1 6794049 95 + 23771897 G-----GCGAUUUAUGCGAUUGCCGCUCGCCAAGUGGCAUAGUGGCUGC-CACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCU------GCUAGAUACUUUA------- (-----((((((.....)))))))(((((((((((((((.......)))-)))))).(....)..))))))....((((((......)------)))))........------- ( -39.30) >DroVir_CAF1 80702 94 + 1 ------CGUAUUUAUGCGAUUGCCGCACCGCAAGUGGCAUAGUGGCUGC-CACUUGAGGAAACACGCGAACAAAAUUAGCGUGCCGCCU------UGCAAUAUUUUA------- ------((((....))))(((((.((.((.(((((((((.......)))-)))))).))...(((((...........)))))..))..------.)))))......------- ( -32.10) >DroGri_CAF1 67550 103 + 1 C-----UGUAUUUAUGCGAUUGCCGCGCCGCAAGUGGCAUAGUGGCUGCCCACUUGAGGAAACACGCGAACAAAAUUAGCGUGCCGCCU------UGCUAUAUUUUAUAGUGCU (-----((((..(((((...((((((.......)))))).(((((....))))).((((...(((((...........)))))...)))------))).)))...))))).... ( -30.70) >DroWil_CAF1 88110 101 + 1 G-----AGUAUUUAUGCGAUUGCCGCCCGGCAAGUGGCAUAGUGACUGC-CACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCUGUUUUGCUAGAUAUUUUA------- (-----(((((((.((((...(((....)))((((((((.......)))-)))))..(....).))))((((((((..(((...)))..))))))))))))))))..------- ( -36.20) >DroMoj_CAF1 73591 100 + 1 CGAGCUCGUAUUUAUGCGAUUGCCGCGCCGCAAGUGGCAUAGUGGCUGC-CACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCU------UGCAAUAUUUUA------- ...((((((......(((.....))).((.(((((((((.......)))-)))))).))......))))))(((((..(((........------)))...))))).------- ( -33.60) >DroAna_CAF1 65694 101 + 1 G-----GGUAUUUAUGCGAUUGCCGCUCGGCAAGUGGCAUAGUGACUGC-CACUUGAGGAAACACGCGAGCCAAAUUAGCGUGCCGCCUUUACGGCUAGAUACUUUA------- (-----(((((((((((((((...(((((.(((((((((.......)))-)))))).(....)...)))))..)))).))))((((......)))).))))))))..------- ( -38.90) >consensus G_____CGUAUUUAUGCGAUUGCCGCGCCGCAAGUGGCAUAGUGGCUGC_CACUUGAGGAAACACGCGAGCAAAAUUAGCGUGCCGCCU______CGAGAUAUUUUA_______ ...............(((.....)))....(((((((((.......))).))))))(((...(((((...........)))))...)))......................... (-22.79 = -22.82 + 0.03)
| Location | 6,794,049 – 6,794,144 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -24.67 |
| Energy contribution | -25.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
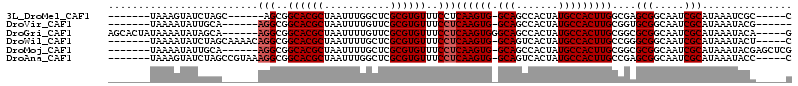
>3L_DroMel_CAF1 6794049 95 - 23771897 -------UAAAGUAUCUAGC------AGCGGCACGCUAAUUUGGCUCGCGUGUUUCCUCAAGUG-GCAGCCACUAUGCCACUUGGCGAGCGGCAAUCGCAUAAAUCGC-----C -------.............------.(((((((((...........)))))).((.(((((((-(((.......)))))))))).))(((.....)))......)))-----. ( -30.60) >DroVir_CAF1 80702 94 - 1 -------UAAAAUAUUGCA------AGGCGGCACGCUAAUUUUGUUCGCGUGUUUCCUCAAGUG-GCAGCCACUAUGCCACUUGCGGUGCGGCAAUCGCAUAAAUACG------ -------........(((.------(((..((((((.(((...))).))))))..))).(((((-(((.......)))))))))))(((((.....))))).......------ ( -32.50) >DroGri_CAF1 67550 103 - 1 AGCACUAUAAAAUAUAGCA------AGGCGGCACGCUAAUUUUGUUCGCGUGUUUCCUCAAGUGGGCAGCCACUAUGCCACUUGCGGCGCGGCAAUCGCAUAAAUACA-----G .((.(((((...)))))..------(((..((((((.(((...))).))))))..)))(((((((.((.......)))))))))..))(((.....))).........-----. ( -28.80) >DroWil_CAF1 88110 101 - 1 -------UAAAAUAUCUAGCAAAACAGGCGGCACGCUAAUUUUGCUCGCGUGUUUCCUCAAGUG-GCAGUCACUAUGCCACUUGCCGGGCGGCAAUCGCAUAAAUACU-----C -------..........(((((((..((((...))))..))))))).((((((..(((((((((-(((.......)))))))))..)))..)))..))).........-----. ( -34.30) >DroMoj_CAF1 73591 100 - 1 -------UAAAAUAUUGCA------AGGCGGCACGCUAAUUUUGCUCGCGUGUUUCCUCAAGUG-GCAGCCACUAUGCCACUUGCGGCGCGGCAAUCGCAUAAAUACGAGCUCG -------.......((((.------..(((((((((...........))))))..((.((((((-(((.......))))))))).))))).))))(((........)))..... ( -33.40) >DroAna_CAF1 65694 101 - 1 -------UAAAGUAUCUAGCCGUAAAGGCGGCACGCUAAUUUGGCUCGCGUGUUUCCUCAAGUG-GCAGUCACUAUGCCACUUGCCGAGCGGCAAUCGCAUAAAUACC-----C -------...........(((((..(((..((((((...........))))))..)))((((((-(((.......)))))))))....)))))...............-----. ( -35.90) >consensus _______UAAAAUAUCGAA______AGGCGGCACGCUAAUUUUGCUCGCGUGUUUCCUCAAGUG_GCAGCCACUAUGCCACUUGCCGAGCGGCAAUCGCAUAAAUACC_____C .........................(((..((((((...........))))))..)))((((((.(((.......)))))))))....(((.....)))............... (-24.67 = -25.00 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:54 2006