
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,789,109 – 6,789,268 |
| Length | 159 |
| Max. P | 0.791357 |

| Location | 6,789,109 – 6,789,205 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.58 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -17.10 |
| Energy contribution | -19.55 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6789109 96 + 23771897 UGCCUGAAUGCAUAAAAUAUUUCUUGUUGUGUGCUUUUGGGCGCCAUUUUGAAGCUGC---CUGGAAAAAUGGCGCUGGGU--GGAAAAUGGUGCUAAAAG .(((..((.(((((.((((.....)))).))))).))..)))((((((((....(..(---(..(..........)..)).--.)))))))))........ ( -31.50) >DroPse_CAF1 62748 96 + 1 CCC-----CCCAAAAAACAUUUCCUGUUGUAUGCUUUUGGGCGCCAUUUUGAAGCUGCAAACUGGAAUAGGGGCUUGGGGUUUGGGGUUUGGGGUUUGGGG (((-----((((((...((..(((((......(((((((....(((.((((......)))).)))..))))))).)))))..))...)))))))...)).. ( -28.90) >DroSec_CAF1 49857 96 + 1 UGCCUGAAUGCAUAAAAUAUUUCUUGUUGUGUGCUUUUGAGCGCCAUUUUGAAGCUGC---CUGGAAAAAUGGCGCCGGGU--GGAAAAUGGUGCUAAAAG .....(((.(((((.((((.....)))).))))).))).(((((((((((....(..(---((((..........))))).--.))))))))))))..... ( -30.90) >DroEre_CAF1 63079 96 + 1 UGGCUGAAUGCAUAAAAUAUUUCUUGUUGUGUGCUUUUGGGCGCCAUUUUGAAGCUGC---CUGGAAAAAUGGCGUUGGGU--GGAAAAUGGGGCGGAAAG ...(..((.(((((.((((.....)))).))))).))..)((.(((((((....(..(---(..(..........)..)).--.)))))))).))...... ( -26.70) >DroYak_CAF1 53637 96 + 1 UGCCUGAAUGCAUAAAAUAUUUCUUGUUGUGUGCUUUUGGGCGCCAUUUUGAAGUUGC---CUGGAAAAAUGGCGUUGGGU--GGAAAAUCGGGCUAAAAG .(((..((.(((((.((((.....)))).))))).))..)))((((((((..((....---))...))))))))(((.(((--.....))).)))...... ( -31.50) >DroAna_CAF1 60403 91 + 1 UGCCUGCCUGCAUAAAAUAUUUCUUGUUGUGUGCUUUUGGGCGCCAUUUUGAAGCUGG--GCUGAAGUCCUGGCUGGGGGC--GGUGGGUGGUG------G .(((..((.(((((.((((.....)))).)))))......((.((.......(((..(--((....).))..))).)).))--))..)))....------. ( -32.00) >consensus UGCCUGAAUGCAUAAAAUAUUUCUUGUUGUGUGCUUUUGGGCGCCAUUUUGAAGCUGC___CUGGAAAAAUGGCGUUGGGU__GGAAAAUGGGGCUAAAAG .(((((((.(((((.((((.....)))).))))).)))))))((((((((..((.......))...))))))))........................... (-17.10 = -19.55 + 2.45)

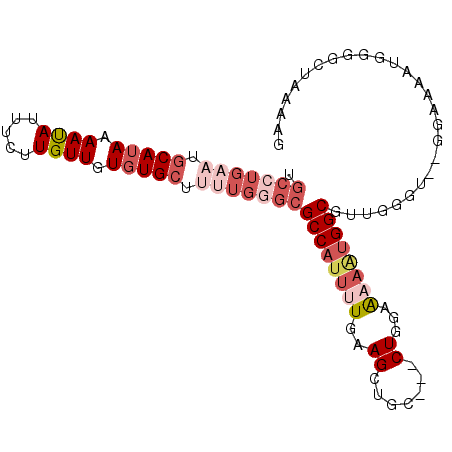
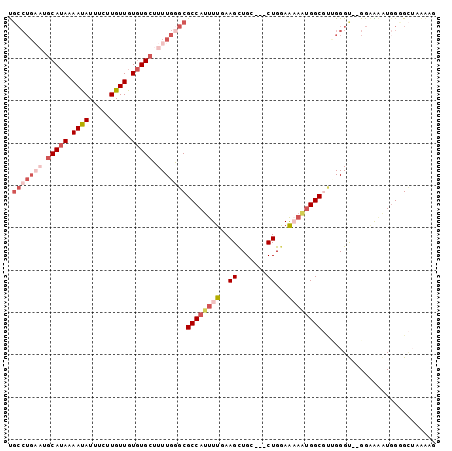
| Location | 6,789,170 – 6,789,268 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.53 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -5.79 |
| Energy contribution | -6.57 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6789170 98 - 23771897 UCAG-GCAACUUCACC---AA-AAAGU-----GCAUCACGCAAUGGCCACUACGGCAUGCUA-UGCACCACCCA------CUUUUAGCACCAUUUUCC--ACCCAGCGCCAUUUUUC ...(-((.........---..-...((-----((((...(((...(((.....))).))).)-)))))......------......((..........--.....)))))....... ( -20.16) >DroPse_CAF1 62807 108 - 1 UCAC-UCAACUUUUACUGCAA-GAACUCUGCGGCCACACGCUAUAG-------GGGGUGUUGUUGCAACACCACCCCGAACCCCAAACCCCAAACCCCAAACCCCAAGCCCCUAUUC ....-............((..-.......(((......)))....(-------(((((((((...))))))).)))...............................))........ ( -19.70) >DroSec_CAF1 49918 99 - 1 UCAG-GCAACUUCACC---AAAAAAGU-----GCAUCACGCAAUUGCCACCACGGCAUGCUA-UGCACCACCCA------CUUUUAGCACCAUUUUCC--ACCCGGCGCCAUUUUUC ...(-((.........---......((-----((((...(((..((((.....))))))).)-)))))......------......((..........--.....)))))....... ( -20.66) >DroSim_CAF1 52493 99 - 1 UCAG-GCAACUUCACC---AAAAAAGU-----GCAUCACGCAAUGGCCACAACGGCAUGCUA-UGCACCACCCA------CUUUUAGCACCAUUUUCC--ACCCGGCGCCAUUUUUC ...(-((.........---......((-----((((...(((...(((.....))).))).)-)))))......------......((..........--.....)))))....... ( -20.06) >DroYak_CAF1 53698 99 - 1 UCAGCACAACUUCACC---GG-AAAGU-----GCAUCACGCAAUGGCCACCACGGCAUGCUA-UGCACCACCCA------CUUUUAGCCCGAUUUUCC--ACCCAACGCCAUUUUUC ................---((-(((((-----((((...(((...(((.....))).))).)-)))))......------.............)))))--................. ( -16.70) >DroAna_CAF1 60465 92 - 1 UCAA-GCAGCUCCACC---AA-AAAGU-----GCAGCACGCAAUGGCCGAGCUAGCAUGUCA-UGCAUCACCAC------C------CACCACCCACC--GCCCCCAGCCAGGACUU ....-(((((((..((---(.-....(-----((.....))).)))..))))).))..(((.-.((........------.------...........--.......))...))).. ( -16.73) >consensus UCAG_GCAACUUCACC___AA_AAAGU_____GCAUCACGCAAUGGCCACCACGGCAUGCUA_UGCACCACCCA______CUUUUAGCACCAUUUUCC__ACCCAGCGCCAUUUUUC ................................(((....(((...(((.....))).)))...)))................................................... ( -5.79 = -6.57 + 0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:52 2006