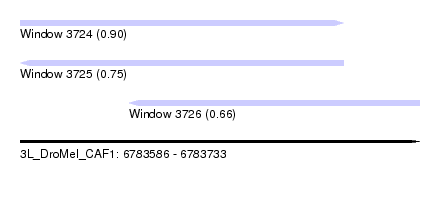
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,783,586 – 6,783,733 |
| Length | 147 |
| Max. P | 0.897076 |

| Location | 6,783,586 – 6,783,705 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -24.32 |
| Energy contribution | -24.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6783586 119 + 23771897 UUUUUAGCCCAAAUUUUCGACAUUGAAUUUCCAGGAUCGAACUUUAAUUUCAGUGUGGUCCGCGGCACAACAUAUUUGCGCUGUGGCACCGUGGUCAACCAGGGCACUUACGCAGUGCC ......((((.........((((((((.((..((.......))..)).))))))))((((((((((.(((.....))).))))))).))).(((....))))))).............. ( -37.00) >DroSec_CAF1 44363 105 + 1 UUUUAAACCCAAAUUUUC--------------GGUAUCGAACUUUAAUCUCAGUGUGGUCCGCGGCACAACAUAUUUGCGCUGUGGCACCGUGGUCAACCAGGGCACUUACGCAGUGCC ......((((.....(((--------------(....))))...............((((((((((.(((.....))).))))))).)))).))).......((((((.....)))))) ( -29.30) >DroSim_CAF1 47917 119 + 1 UUUUAAACCAAAAUUUUCGAGAUUGAAUUUUUGGUAUCGAACUUUAAUCUCAGUGUGGUCCGCGGCACAACAUAUUUGCGCUGUGGCACGGUGGUCAACCAGGGCACUUACGCAGUGCC ......(((.........(((((((((..((((....)))).)))))))))...(((..(((((((.(((.....))).)))))))))))))((....))..((((((.....)))))) ( -37.20) >DroEre_CAF1 57687 113 + 1 UUUGUAGACCAGCACUUUGAAA-UGAUUUCCUA--AUC---AUUUAAUCUCAGUGUGGUCCGCGGCACAACAUAUUUGCGCUGUGGUCCCGUGGUCAACCAGGGCACUUACGCAGUGCC ......((((((((((..((((-(((((....)--)))---)))...))..)))))((.(((((((.(((.....))).))))))).))..)))))......((((((.....)))))) ( -42.40) >DroYak_CAF1 47963 113 + 1 UUUGUCGACUACCAUUUUGAAA-UGAGUGUUCA--AUG---AUUUAAUCUCAGUGUGGUCAGCGGCACAACAUAUUUGCGCUGUGGUCCCGUGGUCAACCGGGGCACUUACGCAGUGCC ..(((((.(((((((.((((..-(((((.....--...---)))))...)))).))))).)))))))..........((((((((((((((.(.....))))))).....)))))))). ( -37.50) >consensus UUUUUAGCCCAAAUUUUCGAAA_UGAAUUUCCAG_AUCGAACUUUAAUCUCAGUGUGGUCCGCGGCACAACAUAUUUGCGCUGUGGCACCGUGGUCAACCAGGGCACUUACGCAGUGCC ....................................................((((.(....).)))).........((((((((...((.(((....))).))......)))))))). (-24.32 = -24.36 + 0.04)

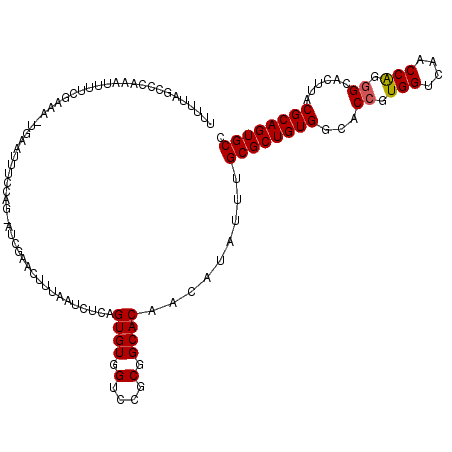
| Location | 6,783,586 – 6,783,705 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -22.56 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6783586 119 - 23771897 GGCACUGCGUAAGUGCCCUGGUUGACCACGGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAAAUUAAAGUUCGAUCCUGGAAAUUCAAUGUCGAAAAUUUGGGCUAAAAA ((((((.....)))))).(((....))).(((.((.(.((((((.....)))))).).)))))...((.(((((....((((((..(((....)))..))))))))))).))....... ( -34.00) >DroSec_CAF1 44363 105 - 1 GGCACUGCGUAAGUGCCCUGGUUGACCACGGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAGUUCGAUACC--------------GAAAAUUUGGGUUUAAAA ((((((.....)))))).(((....))).(((.((.(.((((((.....)))))).).)))))..(((.(((((....((((....)--------------)))))))).)))...... ( -31.40) >DroSim_CAF1 47917 119 - 1 GGCACUGCGUAAGUGCCCUGGUUGACCACCGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAGUUCGAUACCAAAAAUUCAAUCUCGAAAAUUUUGGUUUAAAA ((((((.....))))))......((((((((((.(((((((......))))))).))))).......(((((((.((...............)).))))))).......)))))..... ( -34.46) >DroEre_CAF1 57687 113 - 1 GGCACUGCGUAAGUGCCCUGGUUGACCACGGGACCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAU---GAU--UAGGAAAUCA-UUUCAAAGUGCUGGUCUACAAA (((((.(((((.((((..(((((........)))))..))))...))))).)))))(..(((((((((..((...(((---(((--(....)))))-))))..)))).)))))..)... ( -43.10) >DroYak_CAF1 47963 113 - 1 GGCACUGCGUAAGUGCCCCGGUUGACCACGGGACCACAGCGCAAAUAUGUUGUGCCGCUGACCACACUGAGAUUAAAU---CAU--UGAACACUCA-UUUCAAAAUGGUAGUCGACAAA ((((((.....))))))...(((((((((((...(((((((......)))))))))).))((((...((((((.....---...--.........)-)))))...)))).))))))... ( -32.63) >consensus GGCACUGCGUAAGUGCCCUGGUUGACCACGGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAGUUCGAU_CUAAAAAUUCA_UUUCGAAAAUUUGGGCUAAAAA ((((((.....))))))..(((...((.(((...(((((((......)))))))))).)))))........................................................ (-22.56 = -22.96 + 0.40)

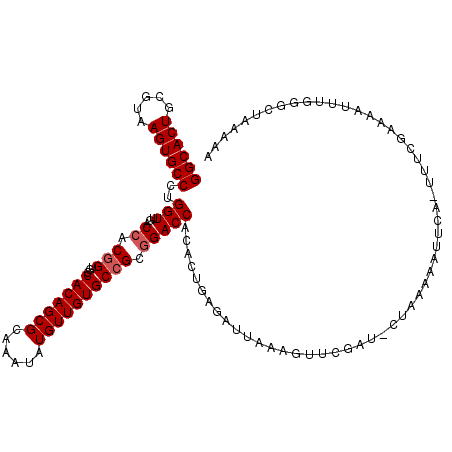
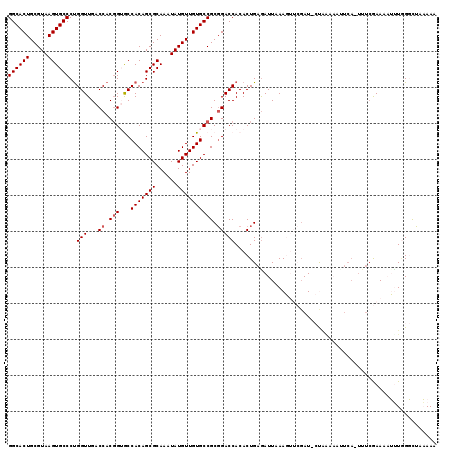
| Location | 6,783,626 – 6,783,733 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.96 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -32.18 |
| Energy contribution | -33.07 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
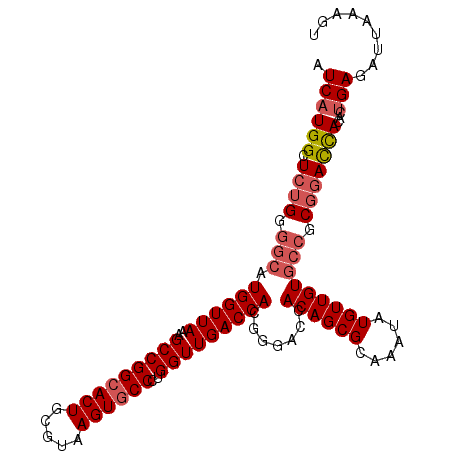
>3L_DroMel_CAF1 6783626 107 - 23771897 AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGCACUGCGUAAGUGCCCUGGUUGACCACGGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAAAUUAAAGU .((((((.((((.((((((((((..(((((((((.....))))))..))))))))...((((....)))).........))))).)))))))...)))......... ( -40.30) >DroSec_CAF1 44389 107 - 1 AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGCACUGCGUAAGUGCCCUGGUUGACCACGGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAGU .((((((.((((.((((((((((..(((((((((.....))))))..))))))))...((((....)))).........))))).)))))))...)))......... ( -40.30) >DroSim_CAF1 47957 107 - 1 AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGCACUGCGUAAGUGCCCUGGUUGACCACCGUGCCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAGU .((((((.((((.((((((((...((((((((((.....))))))..))))....))))))))...((((((.....))))))..)))))))...)))......... ( -41.80) >DroEre_CAF1 57722 106 - 1 AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGCACUGCGUAAGUGCCCUGGUUGACCACGGGACCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAU- .((((((.((((.(((.((((((..(((((((((.....))))))..))))))))).......((((((......))))))))).)))))))...)))........- ( -38.40) >DroYak_CAF1 47998 106 - 1 AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGCACUGCGUAAGUGCCCCGGUUGACCACGGGACCACAGCGCAAAUAUGUUGUGCCGCUGACCACACUGAGAUUAAAU- .((((((((....))).(((((..(((.(((((.(((((.((((((((........))))......))))...))))).))))))))))))))).)))........- ( -36.30) >DroAna_CAF1 54834 107 - 1 AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGGACUGCGUAAGUGCCCUGGUUGACCACGAGACCACAGCGCAAAUAUGUUGUAGGGCGAAUUACACAGAAAGAAAAUU ......(((((((..(.((((((..((((((.(.((....))).)))))))))))).)...))((((((......)))))))))))..................... ( -31.40) >consensus AUCAUGGCUCUGGGGCAUGGUUAAAGCCGGCACUGCGUAAGUGCCCUGGUUGACCACGGGACCACAGCGCAAAUAUGUUGUGCCGCGGACCACACUGAGAUUAAAGU .((((((.((((.(((.((((((..(((((((((.....))))))..))))))))).......((((((......))))))))).)))))))...)))......... (-32.18 = -33.07 + 0.89)

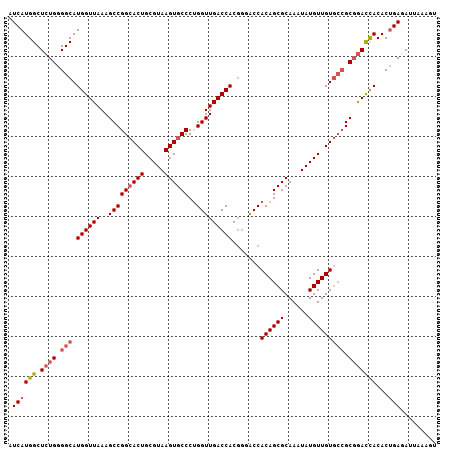
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:45 2006