
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,775,349 – 6,775,471 |
| Length | 122 |
| Max. P | 0.971382 |

| Location | 6,775,349 – 6,775,439 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -24.73 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6775349 90 + 23771897 UA-----CAUACAUAUUUGGGCCAACUUAAAGCGCAGGAAAUUAAAUUUUAUGAUGCCGAGUC--G----------AGUCGAGUCGCAUUGGCAGCU----AAGUUGGCCA ..-----............(((((((((..(((.((.(((((...))))).)).((((((..(--(----------(......)))..)))))))))----))))))))). ( -28.60) >DroSec_CAF1 36186 90 + 1 UAUGGUACAUACAUAUUUGGGCCAACUUAAAGCGCAGGAAAUUAAAUUUUAUGAUGCCGAGUC-----------------GAGUCGCAUUGGCAGCU----AAGUUGGCCA ((((.......))))....(((((((((..(((.((.(((((...))))).)).((((((((.-----------------.....)).)))))))))----))))))))). ( -27.30) >DroSim_CAF1 36614 89 + 1 UAC-AUACAUACAUAUUUGGGCCAACUUAAAGCGCAGGAAAUUAAAUUUUAUGAUGCCGAGUC-----------------GAGUCGCAUUGGCAGCU----AAGUUGGCCA ...-...............(((((((((..(((.((.(((((...))))).)).((((((((.-----------------.....)).)))))))))----))))))))). ( -26.20) >DroYak_CAF1 39721 102 + 1 UA---------CAUAUUUGGGCCAACUUAAAGCGCAGGAAAUUAAAUUUUAUGAUGCCGAGUCGCGAGUCGAGUCGAGUUGAGUCGCAUUGGCAGCUAUGUAAGUUGGCCA ..---------........((((((((((.(((.((.(((((...))))).)).((((((...((((.((((......)))).)))).)))))))))...)))))))))). ( -39.00) >DroAna_CAF1 46418 72 + 1 -------------UAUUUGGGCCAGCUUAAAGCGCAGGAAAUUAAAUUUUAUGAUGCCGAG----------------------UCGCAUUGGCAGCU----AAGUUGGCCA -------------......(((((((((..(((.((.(((((...))))).)).((((((.----------------------.....)))))))))----))))))))). ( -26.40) >DroPer_CAF1 45974 79 + 1 --------GGGCAUAUUUGGGCCAGCUUAAAGUGCAGGAAAUUAAAUUUUAUGAUGCCGAG--------------------AGUCGCAUUGGCAGCU----AAGUUGGCCA --------...........((((((((((.....((.(((((...))))).)).((((((.--------------------.......))))))..)----))))))))). ( -24.30) >consensus UA______AUACAUAUUUGGGCCAACUUAAAGCGCAGGAAAUUAAAUUUUAUGAUGCCGAGUC_________________GAGUCGCAUUGGCAGCU____AAGUUGGCCA ...................(((((((((..(((.((.(((((...))))).)).((((((............................)))))))))....))))))))). (-24.73 = -24.37 + -0.36)

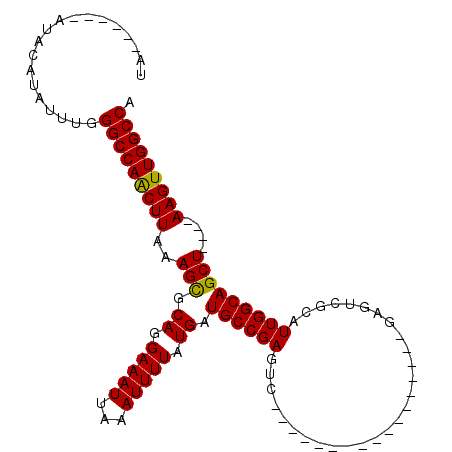
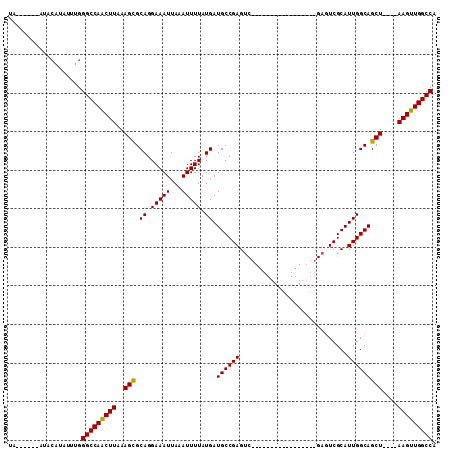
| Location | 6,775,349 – 6,775,439 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -16.62 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6775349 90 - 23771897 UGGCCAACUU----AGCUGCCAAUGCGACUCGACU----------C--GACUCGGCAUCAUAAAAUUUAAUUUCCUGCGCUUUAAGUUGGCCCAAAUAUGUAUG-----UA .(((((((((----((..((.....(((.(((...----------)--)).)))(((..................))))).)))))))))))............-----.. ( -22.77) >DroSec_CAF1 36186 90 - 1 UGGCCAACUU----AGCUGCCAAUGCGACUC-----------------GACUCGGCAUCAUAAAAUUUAAUUUCCUGCGCUUUAAGUUGGCCCAAAUAUGUAUGUACCAUA .(((((((((----((..((.....(((...-----------------...)))(((..................))))).)))))))))))................... ( -20.17) >DroSim_CAF1 36614 89 - 1 UGGCCAACUU----AGCUGCCAAUGCGACUC-----------------GACUCGGCAUCAUAAAAUUUAAUUUCCUGCGCUUUAAGUUGGCCCAAAUAUGUAUGUAU-GUA .(((((((((----((..((.....(((...-----------------...)))(((..................))))).)))))))))))...............-... ( -20.17) >DroYak_CAF1 39721 102 - 1 UGGCCAACUUACAUAGCUGCCAAUGCGACUCAACUCGACUCGACUCGCGACUCGGCAUCAUAAAAUUUAAUUUCCUGCGCUUUAAGUUGGCCCAAAUAUG---------UA .((((((((((...(((((((..(((((.((..........)).)))))....)))).((..((((...))))..)).))).))))))))))........---------.. ( -27.20) >DroAna_CAF1 46418 72 - 1 UGGCCAACUU----AGCUGCCAAUGCGA----------------------CUCGGCAUCAUAAAAUUUAAUUUCCUGCGCUUUAAGCUGGCCCAAAUA------------- .(((((.(((----((..((((((((..----------------------....)))).................)).)).))))).)))))......------------- ( -15.92) >DroPer_CAF1 45974 79 - 1 UGGCCAACUU----AGCUGCCAAUGCGACU--------------------CUCGGCAUCAUAAAAUUUAAUUUCCUGCACUUUAAGCUGGCCCAAAUAUGCCC-------- .(((((.(((----((.(((..((((....--------------------....))))..................)))..))))).)))))...........-------- ( -14.15) >consensus UGGCCAACUU____AGCUGCCAAUGCGACUC_________________GACUCGGCAUCAUAAAAUUUAAUUUCCUGCGCUUUAAGUUGGCCCAAAUAUGUAU______UA .(((((((((....((((((....)))...........................(((..................))))))..)))))))))................... (-16.62 = -17.12 + 0.50)

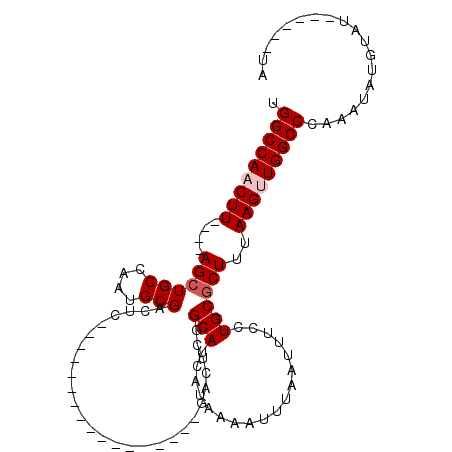
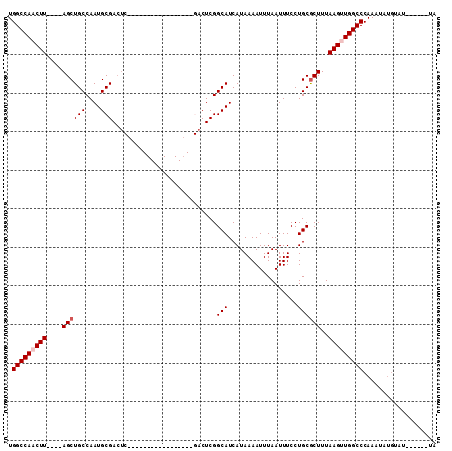
| Location | 6,775,380 – 6,775,471 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -14.03 |
| Energy contribution | -15.03 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6775380 91 + 23771897 GGAAAUUAAAUUUUAUGAUGCCGAGUC--G----------AGUCGAGUCGCAUUGGCAGCU----AAGUUGGCCAUUUGGCCAUUUGGCCAUUCGACCAGGCCCAGA ((..((((.......))))(((..(((--(----------((((((......))))).(((----((((.((((....)))))))))))...)))))..)))))... ( -31.90) >DroSec_CAF1 36222 78 + 1 GGAAAUUAAAUUUUAUGAUGCCGAGUC-----------------GAGUCGCAUUGGCAGCU----AAGUUGGCCAUU--------UGGCCAUUCGACCAGGCCCAGA ((..((((.......))))(((..(((-----------------((((.((..(((((((.----..))).))))..--------..)).)))))))..)))))... ( -23.70) >DroSim_CAF1 36649 86 + 1 GGAAAUUAAAUUUUAUGAUGCCGAGUC-----------------GAGUCGCAUUGGCAGCU----AAGUUGGCCAUUUGGCCAUUUGGCCAUUCGACCAGGCCCAAA ((..((((.......))))(((..(((-----------------((((.((....)).(((----((((.((((....))))))))))).)))))))..)))))... ( -30.20) >DroYak_CAF1 39748 99 + 1 GGAAAUUAAAUUUUAUGAUGCCGAGUCGCGAGUCGAGUCGAGUUGAGUCGCAUUGGCAGCUAUGUAAGUUGGCCAUU--------UGGCCAUUCGACCAGGCCCAGA ((................((((((...((((.((((......)))).)))).))))))(((..((....(((((...--------.)))))....))..)))))... ( -32.50) >DroAna_CAF1 46441 73 + 1 GGAAAUUAAAUUUUAUGAUGCCGAG----------------------UCGCAUUGGCAGCU----AAGUUGGCCAUU--------UGGCCAUUUGGCCAGACCCAGA ((................((((((.----------------------.....))))))(((----((((.((((...--------.))))))))))).....))... ( -24.50) >DroPer_CAF1 46002 72 + 1 GGAAAUUAAAUUUUAUGAUGCCGAG--------------------AGUCGCAUUGGCAGCU----AAGUUGGCCAUU--------UGU---GUUGGCCAGGCACAGG ...............((.((((...--------------------(((.((....)).)))----....((((((..--------...---..)))))))))))).. ( -20.40) >consensus GGAAAUUAAAUUUUAUGAUGCCGAGUC_________________GAGUCGCAUUGGCAGCU____AAGUUGGCCAUU________UGGCCAUUCGACCAGGCCCAGA ((................((((((............................))))))(((........((((((..........))))))........)))))... (-14.03 = -15.03 + 1.00)


| Location | 6,775,380 – 6,775,471 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -14.84 |
| Energy contribution | -16.03 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6775380 91 - 23771897 UCUGGGCCUGGUCGAAUGGCCAAAUGGCCAAAUGGCCAACUU----AGCUGCCAAUGCGACUCGACU----------C--GACUCGGCAUCAUAAAAUUUAAUUUCC ..((((((.((((((.(((((....)))))..((((......----....))))............)----------)--)))).))).)))............... ( -27.60) >DroSec_CAF1 36222 78 - 1 UCUGGGCCUGGUCGAAUGGCCA--------AAUGGCCAACUU----AGCUGCCAAUGCGACUC-----------------GACUCGGCAUCAUAAAAUUUAAUUUCC ..((((((.((((((.(((((.--------...)))))....----.((.......))...))-----------------)))).))).)))............... ( -25.30) >DroSim_CAF1 36649 86 - 1 UUUGGGCCUGGUCGAAUGGCCAAAUGGCCAAAUGGCCAACUU----AGCUGCCAAUGCGACUC-----------------GACUCGGCAUCAUAAAAUUUAAUUUCC ..((((((.((((((.(((((....)))))..((((......----....)))).......))-----------------)))).))).)))............... ( -27.70) >DroYak_CAF1 39748 99 - 1 UCUGGGCCUGGUCGAAUGGCCA--------AAUGGCCAACUUACAUAGCUGCCAAUGCGACUCAACUCGACUCGACUCGCGACUCGGCAUCAUAAAAUUUAAUUUCC ..((((((.((((...(((((.--------...)))))..................((((.((..........)).)))))))).))).)))............... ( -26.30) >DroAna_CAF1 46441 73 - 1 UCUGGGUCUGGCCAAAUGGCCA--------AAUGGCCAACUU----AGCUGCCAAUGCGA----------------------CUCGGCAUCAUAAAAUUUAAUUUCC .((((((((((((....)))))--------..((((......----....))))....))----------------------))))).................... ( -21.50) >DroPer_CAF1 46002 72 - 1 CCUGUGCCUGGCCAAC---ACA--------AAUGGCCAACUU----AGCUGCCAAUGCGACU--------------------CUCGGCAUCAUAAAAUUUAAUUUCC ...(((((((((((..---...--------..))))))....----.((.......))....--------------------...)))))................. ( -17.30) >consensus UCUGGGCCUGGUCGAAUGGCCA________AAUGGCCAACUU____AGCUGCCAAUGCGACUC_________________GACUCGGCAUCAUAAAAUUUAAUUUCC .....(((.((((...((((((..........)))))).........((.......))......................)))).)))................... (-14.84 = -16.03 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:40 2006