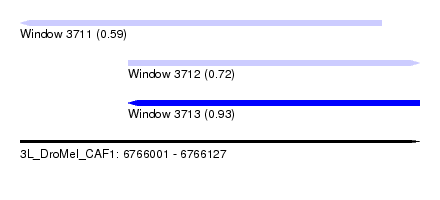
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,766,001 – 6,766,127 |
| Length | 126 |
| Max. P | 0.933980 |

| Location | 6,766,001 – 6,766,115 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -42.73 |
| Consensus MFE | -27.29 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6766001 114 - 23771897 UUGAGGGAUGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGCCAACUGAUGC---GCCGCUACCGCCGAG---GAUUGCAUAUUCUGUGGG .(.((..((.(((((((((((.((((....)))).)))).))))))).))..)).)....((((((..(((........))---)))))))((((..((---(((.....))))))))). ( -45.50) >DroVir_CAF1 40935 96 - 1 UU------GGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGAGCCGCCAAUUGGUGC---GCC---------------GACUGCAUGUUCUGGGGG ..------..(((((((((((.((((....)))).)))).)))))))..(..(.((.(((.(((..((((((....)))).---)).---------------.))))))...)).)..). ( -39.30) >DroGri_CAF1 38020 96 - 1 UU------UGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGCCAACUGAUGC---GCC---------------GACUGCAUGUUCUGGGGA ..------..(((((((((((.((((....)))).)))).)))))))..(..(.((.(((.(((.(((.((........))---)))---------------.))))))...)).)..). ( -39.20) >DroWil_CAF1 53050 120 - 1 UUAUUGGAUGCAGGAGUUGAUCCGGCUGCAGCUGGAUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGUUAACUGAUGCUGCGCCCCCACCGUUGUGGCUGUUUGCAUGUUCUGGGGG .(((..(((.((((((((((((((((....)))))).)))))))))).))...((((.(((....))).)))).)..))).....((((((....((..(....)..)).....)))))) ( -46.20) >DroMoj_CAF1 41185 96 - 1 UU------UGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGAGCCGCCAACUGGUGA---GCC---------------GACUGCAUGUUCUGGGGG ..------..(((((((((((.((((....)))).)))).)))))))..(..(.((.(((.(((..((((((....)))).---)).---------------.))))))...)).)..). ( -39.30) >DroAna_CAF1 36367 111 - 1 UUGAGGGAUGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGCCAACUGGUGC---GCCGCUGCCGCUG------ACUGCAUGUUCUGGGGG .(.((..((.(((((((((((.((((....)))).)))).))))))).))..)).)....((((((((.(((....)))))---.))))))((.(.(------((.....)))..).)). ( -46.90) >consensus UU______UGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGCCAACUGAUGC___GCC_______________GACUGCAUGUUCUGGGGG ..........(((((((((((.((((....)))).)))).)))))))..(..(.((.(((.(((.(((((((....))))....)))................))))))...)).)..). (-27.29 = -27.35 + 0.06)


| Location | 6,766,035 – 6,766,127 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6766035 92 + 23771897 CAUCAGUUGGCGGCCCACUAGCAAUCAGAAAUCCAGGAGUCGAACUCAGCUGCAGCUGCGUCCACUCCUGCAUCCCUCAAUAGUCUAAGCCA .......((((.....((((......((..((.(((((((.((.(.((((....)))).))).))))))).))..))...))))....)))) ( -25.70) >DroVir_CAF1 40957 86 + 1 CACCAAUUGGCGGCUCACUAGCAAUCAGAAAUCCAGGAGUCGAACUCAGCUGCAGCUGCGUCCACUCCUGCC------AACAGUCUGCACCA .......(((.((((....)))...((((....(((((((.((.(.((((....)))).))).)))))))..------.....))))).))) ( -25.00) >DroPse_CAF1 35556 92 + 1 CACCAGUUGGCGGCUCACUAGCAAUCAGAAAUCCAGGAGUCGAACUCAGCUGCAGCUGCGUCCACUCCUGCAUCCCUCAAUAGUCUAAGCCA ..((....)).((((.((((......((..((.(((((((.((.(.((((....)))).))).))))))).))..))...))))...)))). ( -26.80) >DroGri_CAF1 38042 86 + 1 CAUCAGUUGGCGGCCCACUAGCAAUCAGAAAUCCAGGAGUCGAACUCAGCUGCAGCUGCGUCCACUCCUGCA------AAUAGUCUGCACCA .....(((((.......)))))...((((....(((((((.((.(.((((....)))).))).)))))))..------.....))))..... ( -24.30) >DroWil_CAF1 53090 92 + 1 CAUCAGUUAACGGCCCACUAGCAAUCAGAAAUCCAGGAGUCGAAUCCAGCUGCAGCCGGAUCAACUCCUGCAUCCAAUAAUAGUCUGAGUCA ..(((((((...((......))...........(((((((.((.(((.((....)).))))).)))))))..........))).)))).... ( -22.80) >DroPer_CAF1 35195 92 + 1 CACCAGUUGGCGGCUCACUAGCAAUCAGAAAUCCAGGAGUCGAACUCAGCUGCAGCUGCGUCCACUCCUGCAUCCCUCAAUAGUCUAAGCCA ..((....)).((((.((((......((..((.(((((((.((.(.((((....)))).))).))))))).))..))...))))...)))). ( -26.80) >consensus CACCAGUUGGCGGCCCACUAGCAAUCAGAAAUCCAGGAGUCGAACUCAGCUGCAGCUGCGUCCACUCCUGCAUCCCUCAAUAGUCUAAGCCA .......((((.((......))...........(((((((.((.(.((((....)))).))).)))))))..................)))) (-20.94 = -21.05 + 0.11)


| Location | 6,766,035 – 6,766,127 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -30.08 |
| Energy contribution | -29.53 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6766035 92 - 23771897 UGGCUUAGACUAUUGAGGGAUGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGCCAACUGAUG .(((((..((((.(.((..((.(((((((((((.((((....)))).)))).))))))).))..)).)....)))))))))........... ( -38.20) >DroVir_CAF1 40957 86 - 1 UGGUGCAGACUGUU------GGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGAGCCGCCAAUUGGUG (((((((((..(((------..(((((((((((.((((....)))).)))).)))))))))).))))...(((....))))))))....... ( -32.80) >DroPse_CAF1 35556 92 - 1 UGGCUUAGACUAUUGAGGGAUGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGAGCCGCCAACUGGUG .(((((..((((.(.((..((.(((((((((((.((((....)))).)))).))))))).))..)).)....)))))))))(((....))). ( -41.50) >DroGri_CAF1 38042 86 - 1 UGGUGCAGACUAUU------UGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGCCAACUGAUG (((((((((.....------(.(((((((((((.((((....)))).)))).))))))).)..))))...(((....))))))))....... ( -32.80) >DroWil_CAF1 53090 92 - 1 UGACUCAGACUAUUAUUGGAUGCAGGAGUUGAUCCGGCUGCAGCUGGAUUCGACUCCUGGAUUUCUGAUUGCUAGUGGGCCGUUAACUGAUG ....((((.........((((.((((((((((((((((....)))))).)))))))))).)))).((((.(((....))).)))).)))).. ( -36.30) >DroPer_CAF1 35195 92 - 1 UGGCUUAGACUAUUGAGGGAUGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGAGCCGCCAACUGGUG .(((((..((((.(.((..((.(((((((((((.((((....)))).)))).))))))).))..)).)....)))))))))(((....))). ( -41.50) >consensus UGGCUCAGACUAUUGAGGGAUGCAGGAGUGGACGCAGCUGCAGCUGAGUUCGACUCCUGGAUUUCUGAUUGCUAGUGAGCCGCCAACUGAUG ((((.((((.........(((.(((((((((((.((((....)))).)))).))))))).)))))))...(((....))).))))....... (-30.08 = -29.53 + -0.55)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:33 2006