
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,764,387 – 6,764,517 |
| Length | 130 |
| Max. P | 0.891783 |

| Location | 6,764,387 – 6,764,489 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
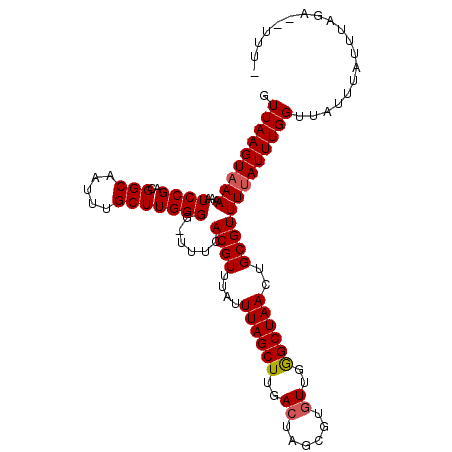
>3L_DroMel_CAF1 6764387 102 - 23771897 GUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGG-UUUCACGUUUAUUUAGCUUGACUAGCG--UUGAGCUAACUGCGUUUUAUUUGGUUAUUUAUUUAGA--UUU- .(((((((((((((((.(.(((.....)))).)))-))).((((....((((((..((....)--)..))))))..))))............))))))))).--...- ( -25.80) >DroSec_CAF1 25323 104 - 1 GUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGG-UUUCACGUUUAUUUAGCUUGACUAGCGUGUUGGGCUAACUGCGUUUUAUUUGGUUAUUUUUUUAGA--UUU- .(((((((((((((((.(.(((.....)))).)))-))).((((....((((((..((......))..))))))..))))))))))))).............--...- ( -25.90) >DroSim_CAF1 25679 104 - 1 GUUAAGUUAAAAAUCCGACGGCAAUUUGCUUGGGG-UUUCACGUUUAUUUAGCUUGACUAGCGUGUUGGGCUAACUGCGUUUUAUUUGGUUAUUUAUUUAGA--UUU- .((((((...((((((.(.(((.....)))).)))-))).((((....((((((..((......))..))))))..))))...............)))))).--...- ( -22.70) >DroYak_CAF1 28362 108 - 1 AUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGUUUCACACGUUUGCUUAGCUUGACUAGCGUGUUGGGCUAACUGCGUUUUAUUUGGUUAUUUAUUUAGAGAUUUU .((((((((((((((((..(((.....))))))))))...((((..(.((((((..((......))..))))))).))))............)))))))))....... ( -26.60) >consensus GUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGG_UUUCACGUUUAUUUAGCUUGACUAGCGUGUUGGGCUAACUGCGUUUUAUUUGGUUAUUUAUUUAGA__UUU_ .(((((((((...((((..(((.....)))))))......((((....((((((..((......))..))))))..)))))))))))))................... (-21.26 = -21.57 + 0.31)

| Location | 6,764,419 – 6,764,517 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6764419 98 - 23771897 GCGUUCAACAAGUUUAUUUCAGCUUUGGGUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGG-UUUCACGUUUAUUUAGCUUGACUAGCG--UUGAGCUA ..(((((((..(.......).((((..(((((((((((((((((.(.(((.....)))).)))-)))....)))))))))))..)..))))--)))))).. ( -29.00) >DroSec_CAF1 25355 100 - 1 GCGUUCAACAAGUUUAUUUCAGCUUUGGGUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGG-UUUCACGUUUAUUUAGCUUGACUAGCGUGUUGGGCUA ..((((((((...........((((..(((((((((((((((((.(.(((.....)))).)))-)))....)))))))))))..)..))).)))))))).. ( -29.10) >DroSim_CAF1 25711 100 - 1 GCGUUCAACAAGUUUAUUUCAGCUUUGGGUUAAGUUAAAAAUCCGACGGCAAUUUGCUUGGGG-UUUCACGUUUAUUUAGCUUGACUAGCGUGUUGGGCUA ((((.(..(((((..(((...((((((((((........))))))).))))))..)))))..)-....)))).....(((((..((......))..))))) ( -27.10) >DroEre_CAF1 38505 98 - 1 GCGCUCAACAAGUUUAUUUCAGCUUUGGGUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGU-UAACACGUUUAUUUAGCUUGACUAGCGUG--GGGCUA ..((((..(((((..(((...((((((((((........))))))).))))))..)))))...-...((((((..((......))..))))))--)))).. ( -25.80) >DroYak_CAF1 28397 101 - 1 GCGCUCAACAAGUUUAUUUCAGCUUUGAAUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGUUUCACACGUUUGCUUAGCUUGACUAGCGUGUUGGGCUA ..((((((((.(((....((((((..(......((...(((((((..(((.....))))))))))...)).....)..))).)))..))).)))))))).. ( -27.30) >consensus GCGUUCAACAAGUUUAUUUCAGCUUUGGGUUAAGUAAAAAAUCCGACGGCAAUUUGCUUGGGG_UUUCACGUUUAUUUAGCUUGACUAGCGUGUUGGGCUA ..((((((((...........((((..(((((((((((...((((..(((.....))))))).........)))))))))))..)..))).)))))))).. (-22.78 = -23.10 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:30 2006