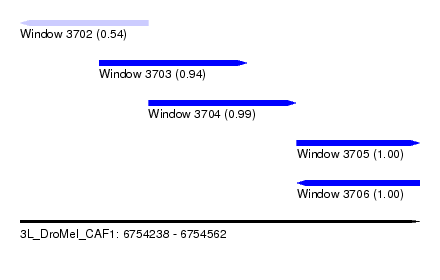
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,754,238 – 6,754,562 |
| Length | 324 |
| Max. P | 0.999932 |

| Location | 6,754,238 – 6,754,342 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -26.35 |
| Consensus MFE | -22.34 |
| Energy contribution | -22.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541220 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6754238 104 - 23771897 UAUGCCCCGAUAUCGGAUCGGCAUCUCCCUCGCUUCUGGCUAGGCAUUCGUAUGUUACAUGGAGAUGUAAGAAUUCCAU--------UUUUGAUUUUAAUCAAUAGAAUCGC-------- .......((((.(((((...((((((((((.((.....)).))((((....)))).....))))))))......)))..--------..(((((....)))))..)))))).-------- ( -24.30) >DroSec_CAF1 17214 104 - 1 UAUGCCCCAAUAUCGGAUCGGCAUCUCCCUCGCUUCUGGCUAGGCAUUCGUAUGUUACAUGGAGAUGUAAGAAUUCCAU--------UUUUGAUUUUAAUCAAUAGAAUCGC-------- ..............(((...((((((((((.((.....)).))((((....)))).....))))))))......)))..--------....(((((((.....)))))))..-------- ( -23.40) >DroSim_CAF1 17544 104 - 1 UAUGCCCCGAUAUCGGAUCGGCAUCUCCCUCGCUUCUGGCUAGGCAUUUGUAUGUUACAUGGAGAUGUAAGAAUUCCAU--------UUUUGAUUUUAAUCAAUAGAAUCGC-------- .......((((.(((((...((((((((((.((.....)).))((((....)))).....))))))))......)))..--------..(((((....)))))..)))))).-------- ( -24.30) >DroEre_CAF1 30128 104 - 1 UAUGCACCGAUAUCGGAUCGGCAUCUCCCUCGCUUCUGGCUAGGCAUUCGUAUGUUACAUGGAGAUGUAAAAAUUCCAU--------UUUUGAUUUUAAUCAAUAGAAUCGA-------- .......((((.(((((...((((((((((.((.....)).))((((....)))).....))))))))......)))..--------..(((((....)))))..)))))).-------- ( -25.00) >DroYak_CAF1 19911 104 - 1 UAUGCCCCGAUAUCGGAUCGGCAUCUCCCUCGCUUCUGGCUAGGCAUUCGUAUGUUACAUGGAGAUGUAAAAAUACCAU--------UUUUGAUUUUAAUCAAUAGAAUCGA-------- ......(((....)))....((((((((((.((.....)).))((((....)))).....))))))))...........--------..(((((((((.....)))))))))-------- ( -24.10) >DroAna_CAF1 26679 119 - 1 UAUGGCCCGGUAUCGGGCCGGCAUCUCCCUCGCUUCUGGCUA-ACAUUCGAAUGUUACAUGGAGAUGUAUAAAUUCCAUUUUUUCAUUUUUGAUUUUAAUCAAUAGAAUCGCAAAUGCCC ..(((((((....)))))))((((((((...((.....))((-((((....))))))...))))))))................(((((.((((((((.....)))))))).)))))... ( -37.00) >consensus UAUGCCCCGAUAUCGGAUCGGCAUCUCCCUCGCUUCUGGCUAGGCAUUCGUAUGUUACAUGGAGAUGUAAAAAUUCCAU________UUUUGAUUUUAAUCAAUAGAAUCGC________ .......((((.(((((...((((((((...((.....))..(((((....)))))....))))))))......)))............(((((....)))))..))))))......... (-22.34 = -22.45 + 0.11)

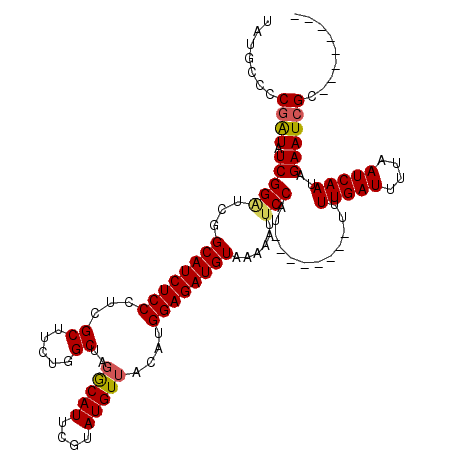
| Location | 6,754,302 – 6,754,422 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.06 |
| Mean single sequence MFE | -35.93 |
| Consensus MFE | -33.88 |
| Energy contribution | -33.97 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6754302 120 + 23771897 GCCAGAAGCGAGGGAGAUGCCGAUCCGAUAUCGGGGCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCGGCCAC ((.....))..(..((((((((((.....)))))((((((....)))))).........)))))..)......(((((...((.(((((((............)))))))))..))))). ( -40.20) >DroSec_CAF1 17278 120 + 1 GCCAGAAGCGAGGGAGAUGCCGAUCCGAUAUUGGGGCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCAC ((.....))..(..((((((((((.....)))))((((((....)))))).........)))))..)......((((....((.(((((((............)))))))))...)))). ( -34.40) >DroSim_CAF1 17608 120 + 1 GCCAGAAGCGAGGGAGAUGCCGAUCCGAUAUCGGGGCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCAC ((.....))..(..((((((((((.....)))))((((((....)))))).........)))))..)......((((....((.(((((((............)))))))))...)))). ( -36.50) >DroEre_CAF1 30192 120 + 1 GCCAGAAGCGAGGGAGAUGCCGAUCCGAUAUCGGUGCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCAC ((.....))..(..(((((..(((.....)))((..(((....)))..)).........)))))..)......((((....((.(((((((............)))))))))...)))). ( -32.00) >DroYak_CAF1 19975 120 + 1 GCCAGAAGCGAGGGAGAUGCCGAUCCGAUAUCGGGGCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCAC ((.....))..(..((((((((((.....)))))((((((....)))))).........)))))..)......((((....((.(((((((............)))))))))...)))). ( -36.50) >DroAna_CAF1 26758 120 + 1 GCCAGAAGCGAGGGAGAUGCCGGCCCGAUACCGGGCCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCAC ((.....))..(..(((((..((((((....))))))........(((......)))..)))))..)......((((....((.(((((((............)))))))))...)))). ( -36.00) >consensus GCCAGAAGCGAGGGAGAUGCCGAUCCGAUAUCGGGGCAUAACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCAC ((.....))..(..((((((((((.....)))))((((((....)))))).........)))))..)......((((....((.(((((((............)))))))))...)))). (-33.88 = -33.97 + 0.08)

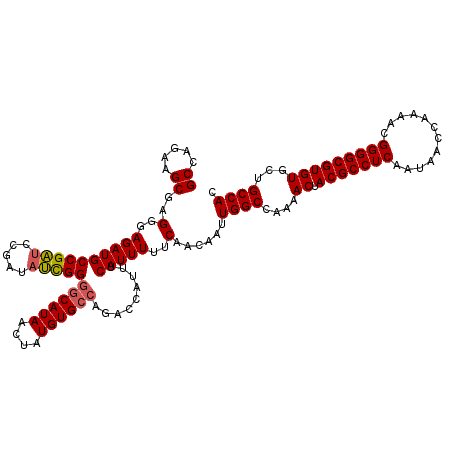
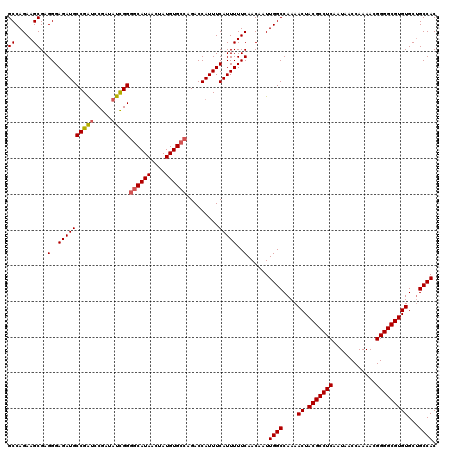
| Location | 6,754,342 – 6,754,462 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.82 |
| Mean single sequence MFE | -31.64 |
| Consensus MFE | -29.93 |
| Energy contribution | -30.10 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6754342 120 + 23771897 ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCGGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCCAGUAAAA (((.((.(((((....................)))))))........................((((((((((.(((((.....((.....))...))))).))))))).)))))).... ( -32.15) >DroSec_CAF1 17318 119 + 1 ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCAAGUAAA- (((..(((((.((.....)).............((((((....((((((((............)))))))).(((((((.........)).))))))))))).).))))....)))...- ( -31.00) >DroSim_CAF1 17648 119 + 1 ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCCAGUAAA- (((..(((((.((.....)).............((((((....((((((((............)))))))).(((((((.........)).))))))))))).).))))....)))...- ( -31.00) >DroEre_CAF1 30232 116 + 1 ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCCAGU---- .....((((..((.....)).............((((((....((((((((............)))))))).(((((((.........)).))))))))))).)))).........---- ( -30.80) >DroYak_CAF1 20015 120 + 1 ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCCAGUCAAA .....((((..((.....)).............((((((....((((((((............)))))))).(((((((.........)).))))))))))).))))............. ( -30.80) >DroAna_CAF1 26798 119 + 1 ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCGAGAGGG- .....((((..((.....)).............((((((....((((((((............)))))))).(((((((.........)).))))))))))).))))...((....)).- ( -34.10) >consensus ACUAUGUGCCAGACCAUUUCAUUUUUCAACAAUUGGCCAAAACUACGCCUCAAUAACCAAAACGGGGCGUGUGCUGCCACAAAAGUAAUGAGCAGUUGGCCACGCGCGCUCCCAGUAAA_ .....((((..((.....)).............((((((....((((((((............)))))))).(((((((.........)).))))))))))).))))............. (-29.93 = -30.10 + 0.17)



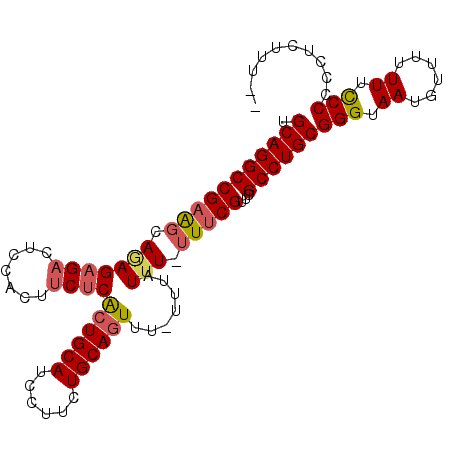
| Location | 6,754,462 – 6,754,562 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -28.36 |
| Energy contribution | -29.12 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -4.34 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6754462 100 + 23771897 AAAAAGAGGGGGGAAAAAAACAUUACCCGCAGGCGAACGAAA-AAAUAAA-AAACUGCAAAAGGAUGCAGUGAGUAGUGGAGUCUCUCUGGUUCGGCCUGCA .......(((.(........)....)))((((((((((....-.......-..((((((......))))))(((.((......)))))..)))).)))))). ( -31.50) >DroSec_CAF1 17437 98 + 1 --AAAGAGGGGGGAAA-AAACAUUACCCGCAGGCGAACGAAA-AAAUAAAAAAACUGCAGAAGGAUGCAUUGAGAAGUGGAGUCUCUCUGCUUCGGCCUGCA --.....(((......-........)))((((((........-............((((......))))....((((..(((...)))..)))).)))))). ( -31.44) >DroSim_CAF1 17767 98 + 1 --AAAGAGGGGGGAAAAAAACAUUACCCGCAGGCGAACGAAA-AAAUAAA-AAACUGCAGAAGGAUGCAGUGAGAAGUGGAGUCUCUCUGCUUCGGCCUGCA --.....(((.(........)....)))((((((........-.......-..((((((......))))))..((((..(((...)))..)))).)))))). ( -36.20) >DroEre_CAF1 30348 98 + 1 --AAAAAGGGGGA-AAAAAACAUUACCCGCAGGCGAACGAAAGAAAUAAA-AAACUGCAUAAGGAUGCAGCGAGAAGUGGAGUCUCUUUGCUACGGCCUGCG --.....(((..(-........)..)))((((((...(....).......-...((((((....))))))...(.((..(((...)))..)).).)))))). ( -29.50) >DroYak_CAF1 20135 99 + 1 UAAAAAAAGGGGG-AAAAAACAUUACCCGCAGGCGAACGAAA-ACAUAAA-AAACUGCAGAAGGAUGCAGCGAGAAGUGGAGUCGCUUUGCCUCGGCCUGCA ........(((..-...........)))((((((........-.......-...(((((......)))))(((((((((....)))))...)))))))))). ( -31.02) >consensus __AAAGAGGGGGGAAAAAAACAUUACCCGCAGGCGAACGAAA_AAAUAAA_AAACUGCAGAAGGAUGCAGUGAGAAGUGGAGUCUCUCUGCUUCGGCCUGCA ........(((..............)))((((((....................(((((......)))))...(((((((((...))))))))).)))))). (-28.36 = -29.12 + 0.76)

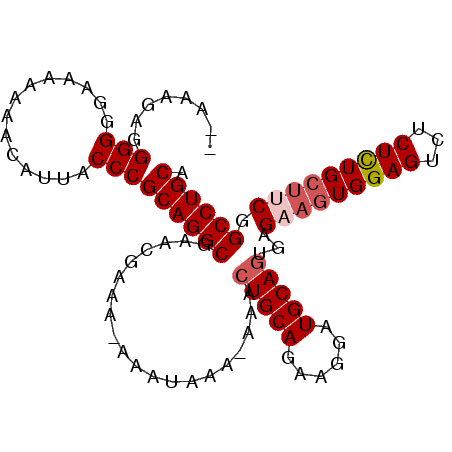
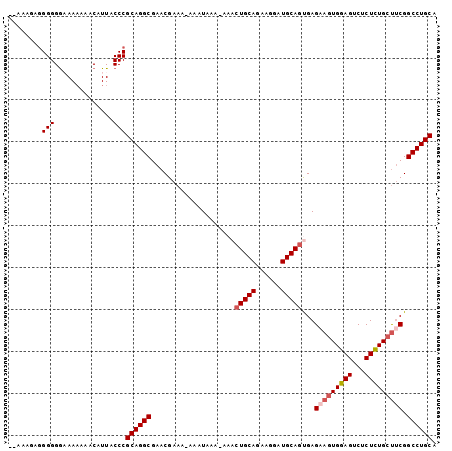
| Location | 6,754,462 – 6,754,562 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6754462 100 - 23771897 UGCAGGCCGAACCAGAGAGACUCCACUACUCACUGCAUCCUUUUGCAGUUU-UUUAUUU-UUUCGUUCGCCUGCGGGUAAUGUUUUUUUCCCCCCUCUUUUU .((((((.((((..(((((......)).)))((((((......))))))..-.......-....))))))))))(((.((......)).))).......... ( -30.10) >DroSec_CAF1 17437 98 - 1 UGCAGGCCGAAGCAGAGAGACUCCACUUCUCAAUGCAUCCUUCUGCAGUUUUUUUAUUU-UUUCGUUCGCCUGCGGGUAAUGUUU-UUUCCCCCCUCUUU-- .(((((((((((..(((((.......)))))..((((......))))............-)))))...))))))(((.((.....-.)).))).......-- ( -25.00) >DroSim_CAF1 17767 98 - 1 UGCAGGCCGAAGCAGAGAGACUCCACUUCUCACUGCAUCCUUCUGCAGUUU-UUUAUUU-UUUCGUUCGCCUGCGGGUAAUGUUUUUUUCCCCCCUCUUU-- .(((((((((((..(((((.......)))))((((((......))))))..-.......-)))))...))))))(((.((......)).)))........-- ( -28.60) >DroEre_CAF1 30348 98 - 1 CGCAGGCCGUAGCAAAGAGACUCCACUUCUCGCUGCAUCCUUAUGCAGUUU-UUUAUUUCUUUCGUUCGCCUGCGGGUAAUGUUUUUU-UCCCCCUUUUU-- .((((((.(.(((...((((.......))))(((((((....)))))))..-............))))))))))(((.((......))-.))).......-- ( -27.90) >DroYak_CAF1 20135 99 - 1 UGCAGGCCGAGGCAAAGCGACUCCACUUCUCGCUGCAUCCUUCUGCAGUUU-UUUAUGU-UUUCGUUCGCCUGCGGGUAAUGUUUUUU-CCCCCUUUUUUUA .((((((.((.(.(((((((.......))..((((((......))))))..-.....))-)))).)).))))))(((.((......))-.)))......... ( -26.20) >consensus UGCAGGCCGAAGCAGAGAGACUCCACUUCUCACUGCAUCCUUCUGCAGUUU_UUUAUUU_UUUCGUUCGCCUGCGGGUAAUGUUUUUUUCCCCCCUCUUU__ .(((((((((((.(((((((.......))))((((((......)))))).......))).)))))...))))))(((.((......)).))).......... (-23.88 = -24.40 + 0.52)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:26 2006