| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 92,764 – 92,862 |
| Length | 98 |
| Max. P | 0.998508 |

| Location | 92,764 – 92,862 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 73.39 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -24.54 |
| Energy contribution | -25.35 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.55 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
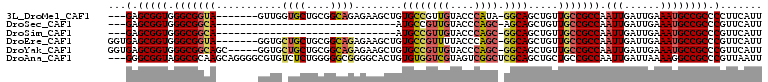
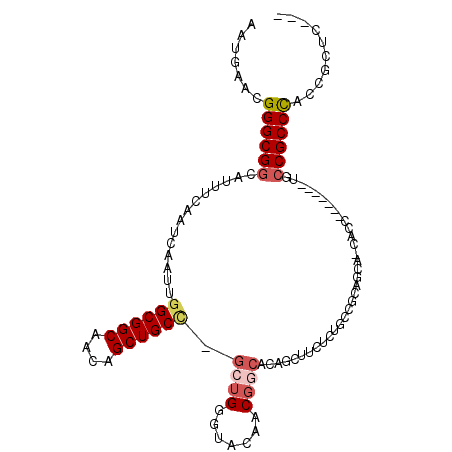
>3L_DroMel_CAF1 92764 98 + 23771897 ---GAGCGGUGGGCGGUA-------GUUGGUGCUGCGGCAGAGAAGCUGUGCCGUUGUACCCAUA-GGCAGCUGUUGCCGCCAAUUGAUUGAAAUGCCGCCCCUUCAUU ---..(.((.((((((((-------...(((((.((((((.((...)).)))))).)))))....-(((.((....)).)))............)))))))))).)... ( -44.80) >DroSec_CAF1 37695 75 + 1 ---GAGCGGUGGGCGGCA------------------------------AUGCCGUUGUACCCAGC-AGCAGCUGUUGCCGCCAAUUGAUUGAAAUGCCGCCCGUUCAUU ---(((((((((((((((------------------------------((((.(((((.....))-))).)).))))))))).(((......)))..)).))))))... ( -33.60) >DroSim_CAF1 42828 75 + 1 ---GAGCGGUGGGCGGCA------------------------------AUGCCGUUGUACCCAGC-GGCAGCUGUUGCCGCCAAUUGAUUGAAAUGCCGCCCGUUCAUU ---(((((((((((((((------------------------------(((((((((....))))-))))....)))))))).(((......)))..)).))))))... ( -36.20) >DroEre_CAF1 56477 101 + 1 GGUGAGCGGUGGGCGGUA-------GGUGCUGCUGCGGCAGAGAAGCUGUGCCGUUUUACCCAGC-GGCAGCUGUUGCCGCCAAUUGAUUGAAAUGCCGCCCGUUCAUU .(((((((((((((((((-------(..((((((((((((.((...)).))))(.....)...))-))))))..)))))))).(((......)))..)).)))))))). ( -47.20) >DroYak_CAF1 53864 103 + 1 GGUGAGCGGUGGGCGGCAGC-----GGUGCUGCUGCGGCAGAGAAGCUGUGCCGUUGUACCCAGC-GGCAGCUGUUGCCGCCAAUUGAUUGAAAUGCCGCCCGUUCAUU .(((((((((((((((((((-----(((((((((((((((.((...)).))))).......))))-)))).))))))))))).(((......)))..)).)))))))). ( -54.91) >DroAna_CAF1 11555 106 + 1 ---GGGCGGUAGGCGCAAGCAGGGGCGUGUCUCUGGGGGCGGGGCACUGUGUGGUCGUAGUCGGCUCGCAGCUGCUGCCGCCAAUUGAUUAAAAGGCCGCCCGUUAAUU ---(((((((..((....))(((((....)))))...((((((((((((((.(((((....)))))))))).)))).))))).............)))))))....... ( -51.80) >consensus ___GAGCGGUGGGCGGCA_______GGUG_UGCUGCGGCAGAGAAGCUGUGCCGUUGUACCCAGC_GGCAGCUGUUGCCGCCAAUUGAUUGAAAUGCCGCCCGUUCAUU ...(.(((((.(((((((...........((((....))))........((((((((....)))).)))).....))))))).(((......)))))))).)....... (-24.54 = -25.35 + 0.81)
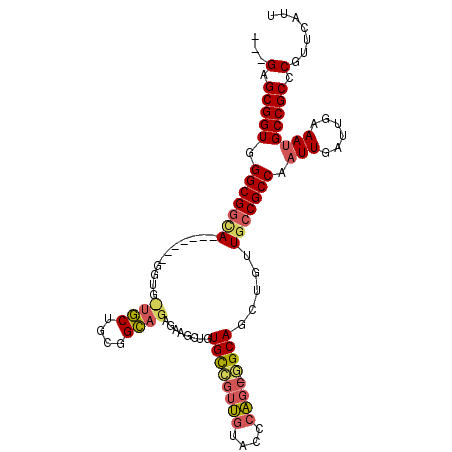
| Location | 92,764 – 92,862 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.39 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -22.92 |
| Energy contribution | -23.67 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 92764 98 - 23771897 AAUGAAGGGGCGGCAUUUCAAUCAAUUGGCGGCAACAGCUGCC-UAUGGGUACAACGGCACAGCUUCUCUGCCGCAGCACCAAC-------UACCGCCCACCGCUC--- .......((((((..............((((((....))))))-....(((.(..(((((.((...)).)))))..).)))...-------..)))))).......--- ( -35.30) >DroSec_CAF1 37695 75 - 1 AAUGAACGGGCGGCAUUUCAAUCAAUUGGCGGCAACAGCUGCU-GCUGGGUACAACGGCAU------------------------------UGCCGCCCACCGCUC--- .......((((((((............((((((....))))))-((((.......))))..------------------------------)))))))).......--- ( -32.70) >DroSim_CAF1 42828 75 - 1 AAUGAACGGGCGGCAUUUCAAUCAAUUGGCGGCAACAGCUGCC-GCUGGGUACAACGGCAU------------------------------UGCCGCCCACCGCUC--- .......((((((((............((((((....))))))-((((.......))))..------------------------------)))))))).......--- ( -35.50) >DroEre_CAF1 56477 101 - 1 AAUGAACGGGCGGCAUUUCAAUCAAUUGGCGGCAACAGCUGCC-GCUGGGUAAAACGGCACAGCUUCUCUGCCGCAGCAGCACC-------UACCGCCCACCGCUCACC .......((((((..............((((((....))))))-((((((((....(((...)))....)))).))))......-------..)))))).......... ( -37.20) >DroYak_CAF1 53864 103 - 1 AAUGAACGGGCGGCAUUUCAAUCAAUUGGCGGCAACAGCUGCC-GCUGGGUACAACGGCACAGCUUCUCUGCCGCAGCAGCACC-----GCUGCCGCCCACCGCUCACC .......(((((((..............((((((..(((((..-((((.......)))).)))))....))))))(((......-----)))))))))).......... ( -43.60) >DroAna_CAF1 11555 106 - 1 AAUUAACGGGCGGCCUUUUAAUCAAUUGGCGGCAGCAGCUGCGAGCCGACUACGACCACACAGUGCCCCGCCCCCAGAGACACGCCCCUGCUUGCGCCUACCGCCC--- .......((((((..............(((((..(((.(((.(.(.((....)).)..).)))))).)))))..........(((........)))....))))))--- ( -29.10) >consensus AAUGAACGGGCGGCAUUUCAAUCAAUUGGCGGCAACAGCUGCC_GCUGGGUACAACGGCACAGCUUCUCUGCCGCAGCA_CACC_______UGCCGCCCACCGCUC___ .......((((((..............((((((....)))))).((((.......))))..................................)))))).......... (-22.92 = -23.67 + 0.75)

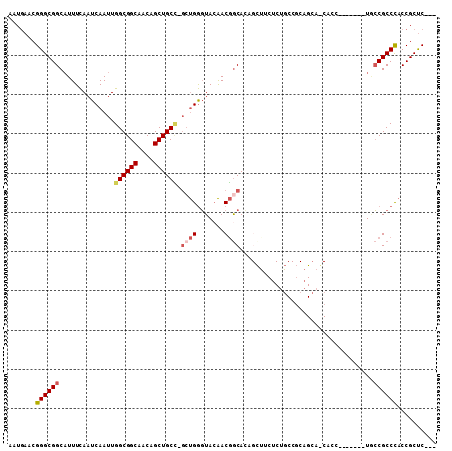
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:09 2006