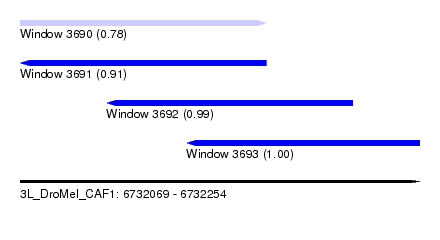
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,732,069 – 6,732,254 |
| Length | 185 |
| Max. P | 0.997444 |

| Location | 6,732,069 – 6,732,183 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -23.73 |
| Energy contribution | -25.60 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6732069 114 + 23771897 CAAAGUGGAGAAGCGGGCAUGGAGUUGCCCAGGUGGGAGCCACCAAUUAGUAAGCCUCGACCCAGUUCAACAAACUGCUCAUGCAGCAAA---AAAGCUGCAGACUUUUUGAAAAGG ....((.(((..(((((((......))))).(((((...))))).........))))).))((..(((((......(.((.((((((...---...)))))))))...)))))..)) ( -37.00) >DroSec_CAF1 4998 116 + 1 CAAAGAGGAGGAGCGAGCAUGGAGUUGCCCAGGUGGGAGCCACCAAUUAGUAAGCCUCGACCCAGUUCAACAAACUGCUCAUGCAGCAAAAAAAAAGCUGCACACUUUUUGAA-AGG ((((((((((...(((((.(((......)))(((((...))))).........).))))...(((((.....)))))))).((((((.........))))))..)))))))..-... ( -34.70) >DroSim_CAF1 3209 116 + 1 CAAAGAGGAGGAGCGAACAUGGAGUUGCCCAGGUGGGAGCCACCAAUUAGUAAGCCUCGACCCAGUUCAACAAACUGCUCAUGCAGCAAAAAAAAAGCUGCACACUUUUUGAA-AGG ((((((((((((((..((.(((......)))(((((...))))).....))..).)))....(((((.....)))))))).((((((.........))))))..)))))))..-... ( -33.90) >DroYak_CAF1 5049 105 + 1 CAAAUGGCA------GGCAAGGAGUUGCCCAGGUGGGAGCCACCAAUUAGUAAGCCUCGACCCAGUUCAACAAACUGCUCAUGCAGAAAA---AUGGAUGCAUACUUGUACAA-A-- .....(((.------(((((....)))))..(((((...))))).........)))........(((((.....((((....))))....---.)))))(((....)))....-.-- ( -25.70) >consensus CAAAGAGGAGGAGCGAGCAUGGAGUUGCCCAGGUGGGAGCCACCAAUUAGUAAGCCUCGACCCAGUUCAACAAACUGCUCAUGCAGCAAA___AAAGCUGCACACUUUUUGAA_AGG (((((((.......(((((.(..((((..(.(((.(..((.((......))..))..).)))..)..))))...)))))).((((((.........))))))..)))))))...... (-23.73 = -25.60 + 1.87)

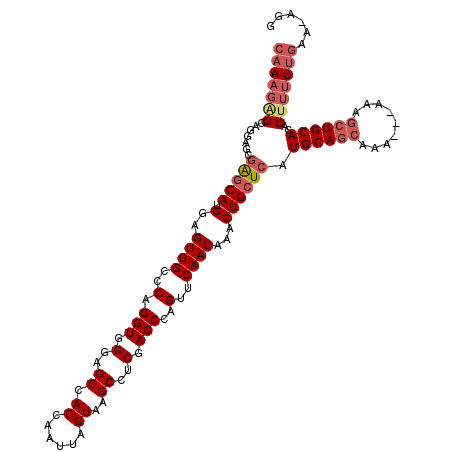
| Location | 6,732,069 – 6,732,183 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -29.21 |
| Energy contribution | -30.40 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6732069 114 - 23771897 CCUUUUCAAAAAGUCUGCAGCUUU---UUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUGGCUCCCACCUGGGCAACUCCAUGCCCGCUUCUCCACUUUG .........(((((.((((((...---...))))))....((((((...))))))((..(((((.........(((((...))))).(((((......)))))))))).))))))). ( -39.10) >DroSec_CAF1 4998 116 - 1 CCU-UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUGGCUCCCACCUGGGCAACUCCAUGCUCGCUCCUCCUCUUUG ...-..((((.((.(((((((.........)))))))(((((....((((..(..(((.(..(((.(((....))))))..).)))..).))))....))))).......)).)))) ( -37.30) >DroSim_CAF1 3209 116 - 1 CCU-UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUGGCUCCCACCUGGGCAACUCCAUGUUCGCUCCUCCUCUUUG ...-..((((.((.(((((((.........)))))))(((((....((((..(..(((.(..(((.(((....))))))..).)))..).))))....))))).......)).)))) ( -34.90) >DroYak_CAF1 5049 105 - 1 --U-UUGUACAAGUAUGCAUCCAU---UUUUCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUGGCUCCCACCUGGGCAACUCCUUGCC------UGCCAUUUG --.-.....(((((.((.(((((.---(((.((((....))))......))).))))))).(((.........(((((...))))).((((((....)))))------))))))))) ( -28.70) >consensus CCU_UUCAAAAAGUGUGCAGCUUU___UUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUGGCUCCCACCUGGGCAACUCCAUGCCCGCUCCUCCUCUUUG ..............(((((((.........)))))))(((((....((((..(..(((.(..(((.(((....))))))..).)))..).))))....))))).............. (-29.21 = -30.40 + 1.19)

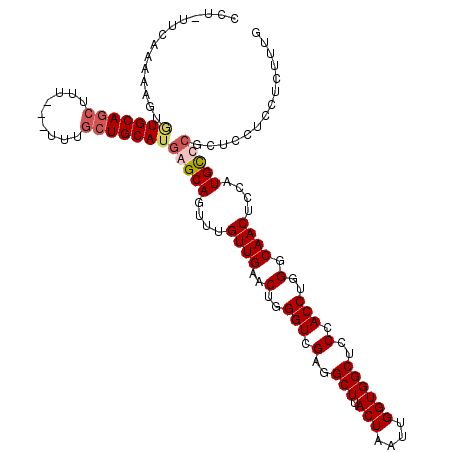
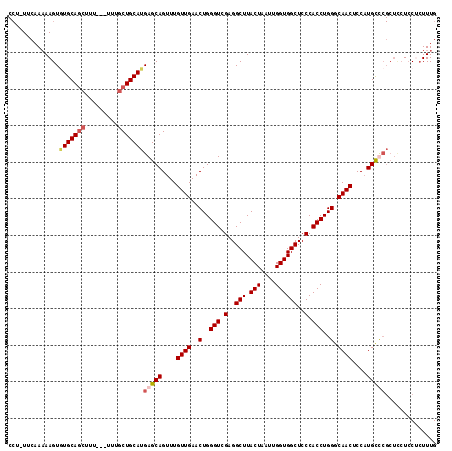
| Location | 6,732,109 – 6,732,223 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.57 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.66 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6732109 114 - 23771897 CCAUUUAUGUAGUAAUCCUUUUAAGUUGCACAUUUACGAACCUUUUCAAAAAGUCUGCAGCUUU---UUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUG (((......((((((.(((..(((((.....)))))(((.((..(((((.(((((((((((...---...)))))).))....))).)))))..)).)))))).))))))..))).. ( -28.10) >DroSec_CAF1 5038 116 - 1 CCAUUUAUGCAGUAUUCCUUUGAAGUUGCACAUUUAAGAACCU-UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUG ((((((((((((((.......((((((((((((((..((....-.))...)))))))))))))).....)))))))))))((((((...))))))(((....))).......))).. ( -36.00) >DroSim_CAF1 3249 116 - 1 CCAUUUAUGCAGUAAUCCUUUUAAGUUGCACAUUUAAGAACCU-UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUG (((((((((((((((.......(((((((((((((..((....-.))...))))))))))))).....))))))))))))((((((...))))))(((....))).......))).. ( -35.60) >consensus CCAUUUAUGCAGUAAUCCUUUUAAGUUGCACAUUUAAGAACCU_UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAGCAGUUUGUUGAACUGGGUCGAGGCUUACUAAUUGGUG (((((((((((((((.......(((((((((((((..(((....)))...))))))))))))).....))))))))))))((((((...))))))(((....))).......))).. (-33.12 = -33.57 + 0.45)

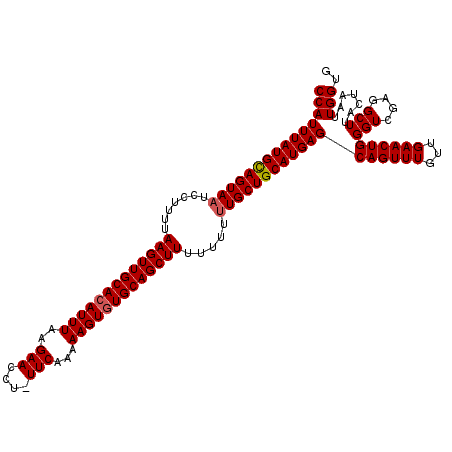

| Location | 6,732,146 – 6,732,254 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.92 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6732146 108 - 23771897 ---------ACAUAGAUAUCAUAUUUAUUAGAGAACUUAACCAUUUAUGUAGUAAUCCUUUUAAGUUGCACAUUUACGAACCUUUUCAAAAAGUCUGCAGCUUU---UUUGCUGCAUGAG ---------..(((((((...)))))))................(((((((((((.......((((((((.((((..(((....)))...)))).)))))))).---.))))))))))). ( -21.30) >DroSec_CAF1 5075 119 - 1 GAACUUAAAACAUAGAUAUAAUAUUUACUAGAGAACUUAACCAUUUAUGCAGUAUUCCUUUGAAGUUGCACAUUUAAGAACCU-UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAG ............................................((((((((((.......((((((((((((((..((....-.))...)))))))))))))).....)))))))))). ( -29.70) >DroSim_CAF1 3286 119 - 1 GAACUUAAAACAUAGAUAUAAUAUUUACUAGAGAACUUAACCAUUUAUGCAGUAAUCCUUUUAAGUUGCACAUUUAAGAACCU-UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAG ............................................(((((((((((.......(((((((((((((..((....-.))...))))))))))))).....))))))))))). ( -29.30) >consensus GAACUUAAAACAUAGAUAUAAUAUUUACUAGAGAACUUAACCAUUUAUGCAGUAAUCCUUUUAAGUUGCACAUUUAAGAACCU_UUCAAAAAGUGUGCAGCUUUUUUUUUGCUGCAUGAG ............................................(((((((((((.......(((((((((((((..(((....)))...))))))))))))).....))))))))))). (-26.82 = -27.27 + 0.45)

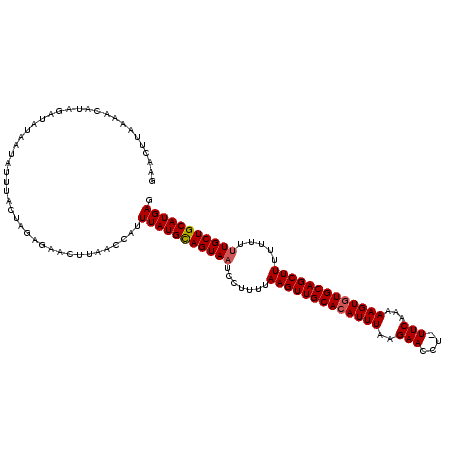
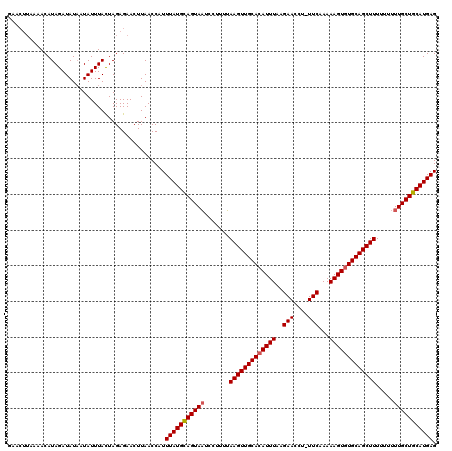
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:14 2006