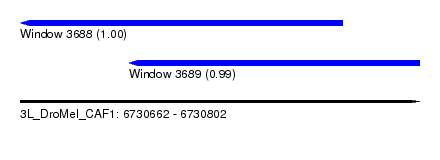
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,730,662 – 6,730,802 |
| Length | 140 |
| Max. P | 0.996236 |

| Location | 6,730,662 – 6,730,775 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.85 |
| Mean single sequence MFE | -39.87 |
| Consensus MFE | -23.10 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6730662 113 - 23771897 GCAACUUUGUGGUUGUGUCUGGUGAAGAACUCAAUUCGCCGCAGGAUUCGAAUCGA---GCC--UGGGCGCCAAACACAAUUGCAUAGCGGCGG--CGGCGAAAUGGCAUCGAAAAAAUG ((..((.((..(((((((.(((((((........)))(((.((((.((((...)))---)))--))))))))).)))))))..)).))..))..--..((......))............ ( -41.30) >DroPse_CAF1 3026 85 - 1 GCCGUUUUGCGGUUGUGUCUGGCGGUUG-----UGCUGCCUCUGGUAUCGCGGC-A---G-C--CGGCUGCCAAACACAAUUGCAGUGCAGCGG-----UGA------------------ ((((((((((((((((((.(((((((((-----.((((((.(.......).)))-)---)-)--))))))))).))))))))))))...)))))-----)..------------------ ( -50.50) >DroSec_CAF1 3607 113 - 1 GCAACUUUGUUGUUGUGUCUGGUGAAGAUCAAAAUUCGCCGCAGGAUUCGAAUCGA---GCC--UGGGCGCCAAACACAAUUGCAUAGCGGCGG--CGGCGAAAUGGCAUCGAAAAAAUG ((..((.(((.(((((((.(((((((........)))(((.((((.((((...)))---)))--))))))))).))))))).))).))..))..--..((......))............ ( -38.20) >DroSim_CAF1 1819 113 - 1 GCAACUUUGUUGUUGUGUCUGGUGAAGAACAAAAUUCGCCGCAGGAUUCGAAUCGA---GCC--UGGGCGCCAAACACAAUUGCAUAGCGGCGG--CGGCGAAAUGGCAUCGAAAAAAUG ((..((.(((.(((((((.(((((((........)))(((.((((.((((...)))---)))--))))))))).))))))).))).))..))..--..((......))............ ( -38.20) >DroYak_CAF1 3632 100 - 1 GCAACUUUGUUGUUGUGUCUGGCGAAGAACUCAAUUCGCCGCAGGAUUCGAAG--------C--UGGGCGCCAAACACAAUUGCACAGCGGCGG--CGGCGAAAUGGAAUCG-------- ((..((.(((.(((((((.(((((((........)))(((.(((.........--------)--))))))))).))))))).))).))..))..--................-------- ( -32.70) >DroAna_CAF1 3586 107 - 1 GCAACUUUGUUUUUGUGUUUGGCGA---------UUUGCAGAGAGCCUCGAGUUUAUCGGCCGGCGAGCGCCAAACACAACUGCACUGCGGCGGAUUGGCAGAAAGG-AUUG---AAAUG .(((((((((..((((((((((((.---------(((((...(.(((..((.....)))))).)))))))))))))))))..)).((((.(.....).)))).))))-.)))---..... ( -38.30) >consensus GCAACUUUGUUGUUGUGUCUGGCGAAGAAC__AAUUCGCCGCAGGAUUCGAAUCGA___GCC__UGGGCGCCAAACACAAUUGCAUAGCGGCGG__CGGCGAAAUGGCAUCG___AAAUG ((..((.(((..((((((.(((((((........)))(((..........................))))))).))))))..))).))..))............................ (-23.10 = -22.55 + -0.55)

| Location | 6,730,700 – 6,730,802 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.98 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6730700 102 - 23771897 ACUCCUGCUAUUAAAUCA--------AGGUU-----GGCUGCAACUUUGUGGUUGUGUCUGGUGAAGAACUCAAUUCGCCGCAGGAUUCGAAUCGA---GCC--UGGGCGCCAAACACAA ................((--------(((((-----(....))))))))...((((((.(((((((........)))(((.((((.((((...)))---)))--))))))))).)))))) ( -33.30) >DroPse_CAF1 3043 103 - 1 AUAACCGCUAUGAUGG-ACAUGAUCGAACAU---G-CACCGCCGUUUUGCGGUUGUGUCUGGCGGUUG-----UGCUGCCUCUGGUAUCGCGGC-A---G-C--CGGCUGCCAAACACAA ..((((((...(((((-.((((......)))---)-.....)))))..))))))((((.(((((((((-----.((((((.(.......).)))-)---)-)--))))))))).)))).. ( -49.00) >DroSec_CAF1 3645 102 - 1 ACACCUGCUAUUAAAUCA--------AGGUU-----GGCUGCAACUUUGUUGUUGUGUCUGGUGAAGAUCAAAAUUCGCCGCAGGAUUCGAAUCGA---GCC--UGGGCGCCAAACACAA ................((--------(((((-----(....))))))))...((((((.(((((((........)))(((.((((.((((...)))---)))--))))))))).)))))) ( -33.30) >DroSim_CAF1 1857 102 - 1 ACACCUGCUAUUAAAUCA--------AGGUU-----GGCUGCAACUUUGUUGUUGUGUCUGGUGAAGAACAAAAUUCGCCGCAGGAUUCGAAUCGA---GCC--UGGGCGCCAAACACAA ................((--------(((((-----(....))))))))...((((((.(((((((........)))(((.((((.((((...)))---)))--))))))))).)))))) ( -33.30) >DroYak_CAF1 3662 97 - 1 ACACCUGCUAUGAAAUCG--------UGGUU-----GGCUGCAACUUUGUUGUUGUGUCUGGCGAAGAACUCAAUUCGCCGCAGGAUUCGAAG--------C--UGGGCGCCAAACACAA ...((.((((((....))--------)))).-----))..(((((...)))))(((((.(((((((........)))(((.(((.........--------)--))))))))).))))). ( -31.60) >DroAna_CAF1 3622 109 - 1 --UGCCGCUAUAAAAUCGCAGCAUCGAGGAUAAUGCGCCCGCAACUUUGUUUUUGUGUUUGGCGA---------UUUGCAGAGAGCCUCGAGUUUAUCGGCCGGCGAGCGCCAAACACAA --(((.((.........)).)))..(((((((((((....)))...))))))))((((((((((.---------(((((...(.(((..((.....)))))).))))))))))))))).. ( -36.20) >consensus ACACCUGCUAUUAAAUCA________AGGUU_____GGCUGCAACUUUGUUGUUGUGUCUGGCGAAGAAC__AAUUCGCCGCAGGAUUCGAAUCGA___GCC__UGGGCGCCAAACACAA ...(((((............................(((.(((((......)))))))).((((((........)))))))))))................................... (-14.86 = -15.20 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:11 2006