
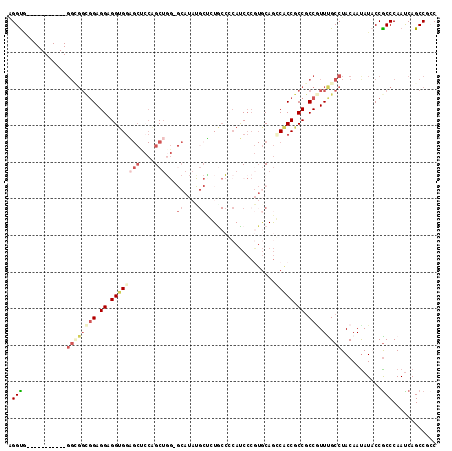
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,729,430 – 6,729,655 |
| Length | 225 |
| Max. P | 0.865307 |

| Location | 6,729,430 – 6,729,538 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.85 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -15.88 |
| Energy contribution | -17.93 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6729430 108 - 23771897 AGGUGGU---------GGCGGCGGAGGAGGUGGAGCUCCAGCUGGUGCAUAUGCUCUACCCCAUCCCGUGCAGGCACCGCCGCCGUUUGCCUACAAUAUACCGCCCAAUCAGCCGCC .((((((---------((((((((.((.((((((((....((....))....))))))))))...((.....))..))))))))((......)).....))))))............ ( -49.30) >DroSec_CAF1 2365 117 - 1 AGGUAGUGGUGGCGGCGGCGGCGGAGGAGGUGGAGCUCCAGCUGGAGCAUAUGCUCUACCCCAUCCCGUGCAGGCACCGCCGCCGUUCGCCUACAACAUACCGCCCAAUCAGCCGCC .((((((.((((((((((((((((.((.((((((((....((....))....))))))))))...((.....))..))))))))).))))).)).)).))))............... ( -58.50) >DroEre_CAF1 2182 105 - 1 CGGC------------GGUGGCGGAGGCGGAGGAGCUCCUGCUGGGGCAUACGCCGUGCCCCAUCCCGUGCAGCCACCGCCGCCGUUCGCCUACAAUAUUCCGCCCAAUCAGCCGGC ((((------------((((((((.(((((((...))))(((((((((((.....))))))))......)))))).))))))))((......)).................)))).. ( -50.70) >DroWil_CAF1 2045 84 - 1 AGGUG------------------GAGGAGGUGGGGCAGCCGC------------CAUG---AAUCCCAUGCAGCCGCCACCGCCGUUUGCCUACAAUAUACCGCCCAAUCAGCCGCC .((((------------------(..((((((((((.((.((------------.(((---.....))))).)).))).)))))((......))..............))..))))) ( -31.40) >DroYak_CAF1 2397 105 - 1 AGGU------------GGCGGCGGAGGAGGUGGAGCCCCAGCUGGCGCAUAUGCUGUGCCGCAUCCCGUGCAGCCACCGCCGCCCUUUGCCUACAAUAUACCACCCAAUCAGCCGGC .(((------------((.((((((((.((((((((....)))(((((((((((......))))...)))).))).)))))..)))))))).........)))))............ ( -42.40) >DroAna_CAF1 2167 90 - 1 GGGUGCU---------GGAGGUGGAGGUGGUGCC------GCUU--------GCGCUGCACCACC----ACAAGCGCCGCCACCAUUUGCCUACAAUAUUCCGCCCAACCAGCCGCC .((((((---------((.(((((((((((((.(------((((--------(.(.((...)).)----.)))))).)))))))..(((....)))...))))))...)))).)))) ( -42.10) >consensus AGGUG___________GGCGGCGGAGGAGGUGGAGCUCCAGCUGG_GCAUAUGCUCUGCCCCAUCCCGUGCAGCCACCGCCGCCGUUUGCCUACAAUAUACCGCCCAAUCAGCCGCC .(((............((((((((.((.((((((((....)))...((....))...................))))).)).))).)))))...........)))............ (-15.88 = -17.93 + 2.06)


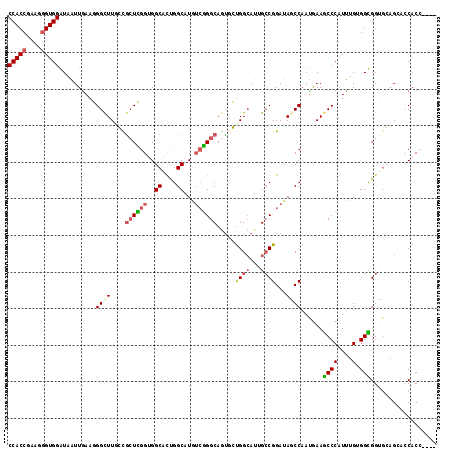
| Location | 6,729,538 – 6,729,655 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -48.87 |
| Consensus MFE | -24.33 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6729538 117 + 23771897 CCACCGAAGGGUGGGUAAUUGAAGGGCUUUGCGCUCGGUGGCAUCGGCAUGUCGGGCAGUGCUGGCAUAGCCGGAUAGCCAAUGAAGCCCAUUUGUGGCGGCGCAGCACCACCGCCG (((((....))))).........(((((((..(((....)))...(((.((((.(((.(((....))).))).)))))))...)))))))......(((((.(......).))))). ( -53.50) >DroGri_CAF1 2351 112 + 1 CCACCGAAAUGUGGAUAAUUAAAUGGCUUGCUACUGGGUGGCGCUGGCAUUUCCGGCAAUUGUGGCAUUGCCGGAUAUCCAAUGAAACCCAUUUGAGGUGGAUGAGGCACAC----- ((((......)))).........((.(((((((((((((..((.(((.((.(((((((((......))))))))).))))).))..))))......))))).)))).))...----- ( -38.90) >DroSec_CAF1 2482 117 + 1 CCACCGAAGGGUGGGUAAUUGAAGGGCUUUGCGCUCGGUGGCAUCGGCAUGUCGGGCAGUGCUGGCAUGGCCGGAUAGCCAAUGAAGCCCAUUUGGGGCGGUGCAGCCCCACCGCCG (((((....)))))....(..(((((((((..(((....)))...(((.((((.(((.(((....))).))).)))))))...))))))).))..)(((((((......))))))). ( -58.20) >DroWil_CAF1 2129 113 + 1 CCACCAAAGGGUGGAUAGUUGAAUGGUUUGCUGGUCGGGGGCUCUGGCAUGGAGGGCAAUGGCGGAACUGCCGGAUAACCAAUGAAACCCAUCUGUGGUGGAGGUGCUCCACC---- (((((....)))))((((.((...(((((..(((((((.((.((((.(((........))).)))).)).)))....))))...))))))).))))(((((((...)))))))---- ( -42.30) >DroYak_CAF1 2502 117 + 1 CCACCGAAGGGUGGGUAAUUGAAGGGCUUGGCGCUCGGUGGCAUUGGCAUAUCGGGCAGUGCUGGCAUAGCCGGAUAACCAACAAAGCCCAUUUGUGGCGGUGCAGGACCACCUCCG (((((....))))).........(((((((((((((((((((....)).)))))))).(((....))).)))((....))....))))))......((.((((......)))).)). ( -49.50) >DroMoj_CAF1 1967 112 + 1 CCACCGAAGGGUGGAUAAUUGAAUGGCUUGCUGCUGGGAGGCGCUGGCAUGUCUGGCAGCUGUGGCAUUGGCGGAUAGCCAAUGAAGCCCAUUUGAGGUGGUGCAGGGGCAC----- (((((....)))))....(..(((((((((((((..((..((....))...))..))))).....(((((((.....)))))))))).)))))..)....((((....))))----- ( -50.80) >consensus CCACCGAAGGGUGGAUAAUUGAAGGGCUUGCCGCUCGGUGGCACUGGCAUGUCGGGCAGUGCUGGCAUUGCCGGAUAGCCAAUGAAGCCCAUUUGUGGCGGUGCAGCACCACC____ (((((....)))))..........((((....((((((..((....))...)))))).....((((...))))...))))......((((....).))).................. (-24.33 = -24.58 + 0.25)


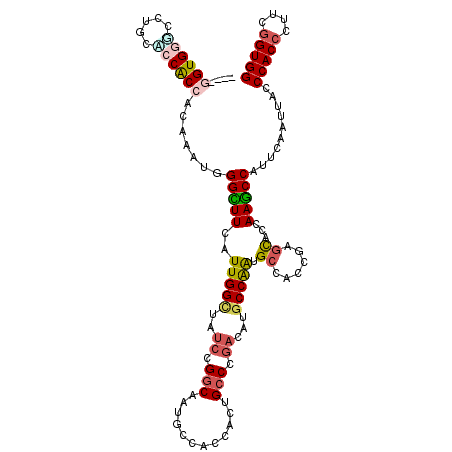
| Location | 6,729,538 – 6,729,655 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.30 |
| Mean single sequence MFE | -40.03 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6729538 117 - 23771897 CGGCGGUGGUGCUGCGCCGCCACAAAUGGGCUUCAUUGGCUAUCCGGCUAUGCCAGCACUGCCCGACAUGCCGAUGCCACCGAGCGCAAAGCCCUUCAAUUACCCACCCUUCGGUGG .(((((((......)))))))......(((((((((((((..((.(((..((....))..))).))...)))))))............)))))).........(((((....))))) ( -46.40) >DroGri_CAF1 2351 112 - 1 -----GUGUGCCUCAUCCACCUCAAAUGGGUUUCAUUGGAUAUCCGGCAAUGCCACAAUUGCCGGAAAUGCCAGCGCCACCCAGUAGCAAGCCAUUUAAUUAUCCACAUUUCGGUGG -----...........(((((.....(((((..(.((((((.(((((((((......))))))))).)).)))).)..)))))((.....))....................))))) ( -33.70) >DroSec_CAF1 2482 117 - 1 CGGCGGUGGGGCUGCACCGCCCCAAAUGGGCUUCAUUGGCUAUCCGGCCAUGCCAGCACUGCCCGACAUGCCGAUGCCACCGAGCGCAAAGCCCUUCAAUUACCCACCCUUCGGUGG .(((..((((((......))))))..((.(((((((((((..((.(((..((....))..))).))...))))))).....)))).))..)))..........(((((....))))) ( -46.30) >DroWil_CAF1 2129 113 - 1 ----GGUGGAGCACCUCCACCACAGAUGGGUUUCAUUGGUUAUCCGGCAGUUCCGCCAUUGCCCUCCAUGCCAGAGCCCCCGACCAGCAAACCAUUCAACUAUCCACCCUUUGGUGG ----(((((((...)))))))...((..(((((..((((((....((((((......))))))(((.......))).....)))))).)))))..))......(((((....))))) ( -36.00) >DroYak_CAF1 2502 117 - 1 CGGAGGUGGUCCUGCACCGCCACAAAUGGGCUUUGUUGGUUAUCCGGCUAUGCCAGCACUGCCCGAUAUGCCAAUGCCACCGAGCGCCAAGCCCUUCAAUUACCCACCCUUCGGUGG .(((((((((.....))))))......((((((.((((((((((.(((..((....))..))).)))).))))))((......))...)))))))))......(((((....))))) ( -42.90) >DroMoj_CAF1 1967 112 - 1 -----GUGCCCCUGCACCACCUCAAAUGGGCUUCAUUGGCUAUCCGCCAAUGCCACAGCUGCCAGACAUGCCAGCGCCUCCCAGCAGCAAGCCAUUCAAUUAUCCACCCUUCGGUGG -----((((....))))...........((((((((((((.....))))))).....(((((..((..((....))..))...))))))))))..........(((((....))))) ( -34.90) >consensus ____GGUGGGCCUGCACCACCACAAAUGGGCUUCAUUGGCUAUCCGGCAAUGCCACCACUGCCCGACAUGCCAAUGCCACCGAGCACCAAGCCAUUCAAUUACCCACCCUUCGGUGG ....((((((.....)))))).......(((((..(((((..((.(((............))).))...))))).((......))...)))))..........(((((....))))) (-24.28 = -24.62 + 0.34)

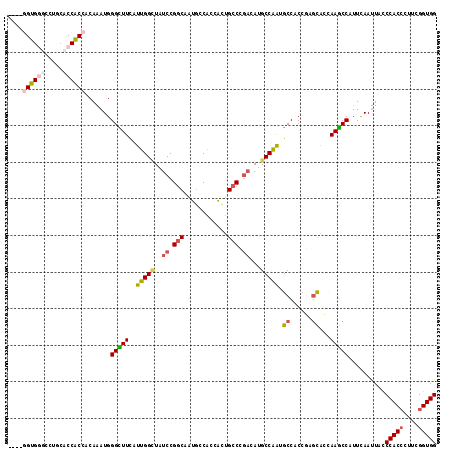
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:09 2006