
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,706,441 – 6,706,568 |
| Length | 127 |
| Max. P | 0.984255 |

| Location | 6,706,441 – 6,706,537 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6706441 96 + 23771897 CAUAAGUUGGUGGCGCAAUUUUCUGCU---G------CUGUUGCUGCAAAUUAUCCACGGCGUCUGCUGCGGCAGCAGCAUUUGCUGCGGCAUUAGUAGACUGAU ......((((((.((((......((((---(------(((((((.(((.((...(....).)).))).)))))))))))).....)))).))))))......... ( -35.20) >DroSec_CAF1 2086 99 + 1 CAUAAGUUGGUGGUGCGAUUUUCUGCUGCUG------CUGUUGCUGCAAAUUAUCCACGGCGUCUGCUGCGGCAGCAGCAUUUGCUGCGGCAUUAGUAGACUGAU .........((.(((.(((..(.(((.((..------.....)).))).)..)))))).))((((((((..((.(((((....))))).))..)))))))).... ( -37.90) >DroYak_CAF1 2122 90 + 1 CAUAAGUUGGAGGCGCAAUUUUCUGCUGCUGUUGCUGUUG---------------CACGGCGUCUGCUGCAGCUGCAGCAUUGGCAGCGGCAUUAGUAGACUGAU ...((((((......))))))((..((((((.((((((((---------------(.(((....((((((....)))))))))))))))))).))))))...)). ( -34.60) >consensus CAUAAGUUGGUGGCGCAAUUUUCUGCUGCUG______CUGUUGCUGCAAAUUAUCCACGGCGUCUGCUGCGGCAGCAGCAUUUGCUGCGGCAUUAGUAGACUGAU .....((((.....(((......)))............(((....))).........))))(((((((((.(((((.......))))).))...))))))).... (-24.35 = -24.43 + 0.09)



| Location | 6,706,441 – 6,706,537 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6706441 96 - 23771897 AUCAGUCUACUAAUGCCGCAGCAAAUGCUGCUGCCGCAGCAGACGCCGUGGAUAAUUUGCAGCAACAG------C---AGCAGAAAAUUGCGCCACCAACUUAUG ....((((.....(((.((((((.....)))))).)))..))))...((((....(((((.((....)------)---.)))))........))))......... ( -27.70) >DroSec_CAF1 2086 99 - 1 AUCAGUCUACUAAUGCCGCAGCAAAUGCUGCUGCCGCAGCAGACGCCGUGGAUAAUUUGCAGCAACAG------CAGCAGCAGAAAAUCGCACCACCAACUUAUG ....((((.....(((.((((((.....)))))).)))..))))...((((......(((.((....)------).)))((........)).))))......... ( -28.80) >DroYak_CAF1 2122 90 - 1 AUCAGUCUACUAAUGCCGCUGCCAAUGCUGCAGCUGCAGCAGACGCCGUG---------------CAACAGCAACAGCAGCAGAAAAUUGCGCCUCCAACUUAUG ....((((.....(((.(((((.......))))).)))..))))((..((---------------(....)))...)).((((....)))).............. ( -22.70) >consensus AUCAGUCUACUAAUGCCGCAGCAAAUGCUGCUGCCGCAGCAGACGCCGUGGAUAAUUUGCAGCAACAG______CAGCAGCAGAAAAUUGCGCCACCAACUUAUG ....((((.....(((.((((((.....)))))).)))..))))...................................((........)).............. (-18.34 = -18.23 + -0.11)


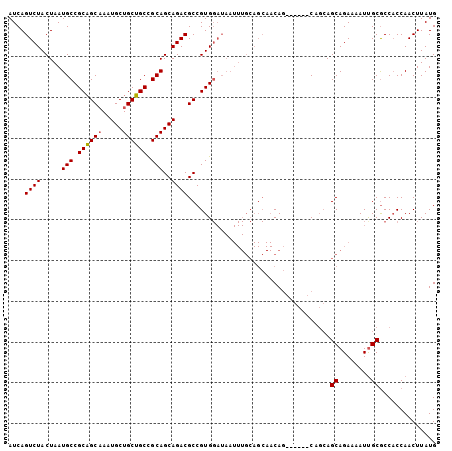
| Location | 6,706,469 – 6,706,568 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6706469 99 + 23771897 CUGUUGCUGCAAAUUAUCCACGGCGUCUGCUGCGGCAGCAGCAUUUGCUGCGGCAUUAGUAGACUGAUACUGCGCAUAGGUUGGAUAUUUCGGUUUGUG .....((((.((((.(((((.((((..((((((....))))))..))))((((.(((((....))))).))))........)))))))))))))..... ( -34.30) >DroSec_CAF1 2117 99 + 1 CUGUUGCUGCAAAUUAUCCACGGCGUCUGCUGCGGCAGCAGCAUUUGCUGCGGCAUUAGUAGACUGAUAUUGCGCAUAGGUUGGAUAUUUCGGUUUGUG .....((((.((((.((((((.((((..((((((((((......))))))))))(((((....)))))...))))....).)))))))))))))..... ( -32.10) >DroEre_CAF1 2170 99 + 1 CUGUUGCUGCAAAUUAUCCACGGCAUCUGCUGCGGCUGCAGCAUUGGCUGCGGCAUUAGUAGACUGAUAUUGCGCAUAGGUUGGAUAUUUCGGUUUGUG .....((((.((((.(((((....(((((.((((((((((((....))))))))(((((....)))))....))))))))))))))))))))))..... ( -35.20) >DroYak_CAF1 2159 84 + 1 UUG---------------CACGGCGUCUGCUGCAGCUGCAGCAUUGGCAGCGGCAUUAGUAGACUGAUAUUGCGCAUAGGUUGGAUAUUUCGGCUUGUG ..(---------------((...(((((((((..(((((.((....)).)))))..)))))))).)....))).(((((((((((...))))))))))) ( -31.30) >consensus CUGUUGCUGCAAAUUAUCCACGGCGUCUGCUGCGGCAGCAGCAUUGGCUGCGGCAUUAGUAGACUGAUAUUGCGCAUAGGUUGGAUAUUUCGGUUUGUG ........................((((((((..((((((((....))))))))..))))))))..........((((((..(((...)))..)))))) (-26.94 = -27.75 + 0.81)


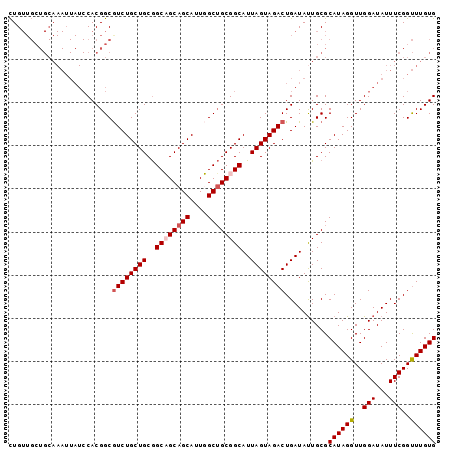
| Location | 6,706,469 – 6,706,568 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.05 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -18.10 |
| Energy contribution | -20.35 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6706469 99 - 23771897 CACAAACCGAAAUAUCCAACCUAUGCGCAGUAUCAGUCUACUAAUGCCGCAGCAAAUGCUGCUGCCGCAGCAGACGCCGUGGAUAAUUUGCAGCAACAG .......((((.((((((.....(((..((((......))))..(((.((((((.....)))))).)))))).......)))))).))))......... ( -27.40) >DroSec_CAF1 2117 99 - 1 CACAAACCGAAAUAUCCAACCUAUGCGCAAUAUCAGUCUACUAAUGCCGCAGCAAAUGCUGCUGCCGCAGCAGACGCCGUGGAUAAUUUGCAGCAACAG .......................(((((((.....((((((...(((.((((((.....)))))).)))((....)).))))))...)))).))).... ( -27.10) >DroEre_CAF1 2170 99 - 1 CACAAACCGAAAUAUCCAACCUAUGCGCAAUAUCAGUCUACUAAUGCCGCAGCCAAUGCUGCAGCCGCAGCAGAUGCCGUGGAUAAUUUGCAGCAACAG .......................(((((((.....((((((....((.(((((....))))).)).(((.....))).))))))...)))).))).... ( -25.20) >DroYak_CAF1 2159 84 - 1 CACAAGCCGAAAUAUCCAACCUAUGCGCAAUAUCAGUCUACUAAUGCCGCUGCCAAUGCUGCAGCUGCAGCAGACGCCGUG---------------CAA .......................(((((.......((((.....(((.(((((.......))))).)))..))))...)))---------------)). ( -20.30) >consensus CACAAACCGAAAUAUCCAACCUAUGCGCAAUAUCAGUCUACUAAUGCCGCAGCAAAUGCUGCAGCCGCAGCAGACGCCGUGGAUAAUUUGCAGCAACAG .......((((.((((((.....((((...(((.((....)).))).))))((...((((((....))))))...))..)))))).))))......... (-18.10 = -20.35 + 2.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:02 2006