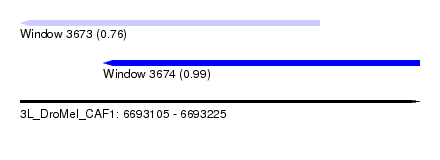
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,693,105 – 6,693,225 |
| Length | 120 |
| Max. P | 0.985396 |

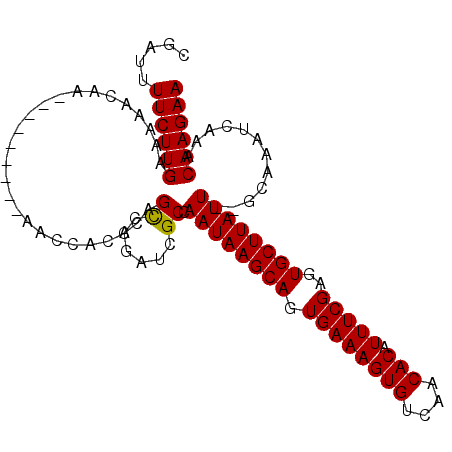
| Location | 6,693,105 – 6,693,195 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -14.49 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6693105 90 - 23771897 CAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--G-CAAAUCAAACAAGAAAAGCGUAACCAAGCACAUA----AGCAAA ..(((...(((((((((.((((((((....))).)))))..))))))))--)-...)))...........((........)).....----...... ( -20.70) >DroPse_CAF1 68113 88 - 1 CCUCGCGCCAAUAAGCAGUGAAAGUGUCAAUACAUUUCGAGUGCUUAUU--GCCAAAUCAAAUACGAUAAACAAA---AAGCCAACA----AGCAAA ....(((.(((((((((.(..((((((....))))))..).))))))))--).)..(((......))).......---.........----.))... ( -18.60) >DroSec_CAF1 69673 90 - 1 CAGAUCGCAAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--G-CAAUUCAAACAAGAAAAGCGUAACCAAGCAUAUA----AGCAAA ......((.((((((((.((((((((....))).)))))..))))))))--(-(..(((......)))..))........)).....----...... ( -19.70) >DroSim_CAF1 88187 90 - 1 CAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--G-CAAAUCAAACAAGAAAAGCGUAACCAAGCAUAUA----AGCAAA ..(((...(((((((((.((((((((....))).)))))..))))))))--)-...)))...........((........)).....----...... ( -20.70) >DroAna_CAF1 91362 95 - 1 C-GAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUUCGC-CAAAUCAAACAAGAAAAGCGCAACCAAGCAAAUAACCAAGCAAA .-(((.((.((((((((.((((((((....))).)))))..)))))))).))-...)))...........((........))............... ( -20.30) >DroPer_CAF1 68244 88 - 1 CCUCGCGCCAAUAAGCAGUGAAAGUGUCAAUACAUUUCGAGUGCUUAUU--GCCAAAUCAAAUACGAUAAACAAA---AAGCCAACA----AGCAAA ....(((.(((((((((.(..((((((....))))))..).))))))))--).)..(((......))).......---.........----.))... ( -18.60) >consensus CAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU__G_CAAAUCAAACAAGAAAAGCGUAACCAAGCAAAUA____AGCAAA ......((.((((((((.((((((((....))).)))))..)))))))).....................((........))..........))... (-14.49 = -14.60 + 0.11)

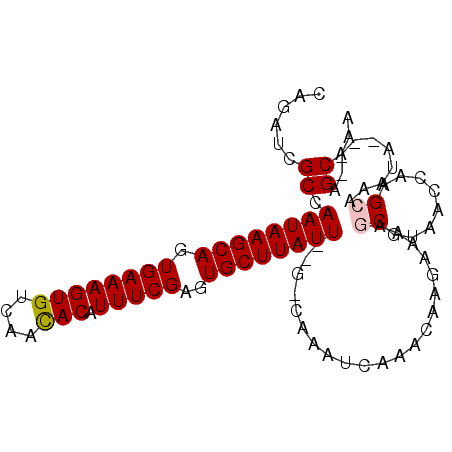
| Location | 6,693,130 – 6,693,225 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.25 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -18.74 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6693130 95 - 23771897 CGAUUUUCUUGAAAAACAA---------AACCACCCAGCCAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--GCAAAUCAAACAAGAA .....((((((........---------.........((......))(((((((((.((((((((....))).)))))..))))))))--).........)))))) ( -21.80) >DroSec_CAF1 69698 95 - 1 CGAUUUUCUUGAAAAACAA---------AACCACCCAGUCAGAUCGCAAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--GCAAUUCAAACAAGAA .....((((((........---------.........(......)((((.((((((.((((((((....))).)))))..))))))))--))........)))))) ( -20.80) >DroSim_CAF1 88212 95 - 1 CGAUUUUCUUGAAAAACAA---------AACCACCCAGCCAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--GCAAAUCAAACAAGAA .....((((((........---------.........((......))(((((((((.((((((((....))).)))))..))))))))--).........)))))) ( -21.80) >DroEre_CAF1 81628 88 - 1 CGAUUUUCUUGAAA----------------ACCCCCAGCCAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--GCAAAUCAAACAAGAA .....((((((...----------------.......((......))(((((((((.((((((((....))).)))))..))))))))--).........)))))) ( -21.80) >DroYak_CAF1 82582 104 - 1 CGAUUUUCUUGAAAAAAAAUCAAAAAAUAAAAUCCCAGCCAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU--GCAAAUCAAACAAGAA .....((((((..........................((......))(((((((((.((((((((....))).)))))..))))))))--).........)))))) ( -21.80) >DroAna_CAF1 91391 91 - 1 CGAAUUUCUUGUCCA--------------GUUCCCCAGCC-GAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUUCGCCAAAUCAAACAAGAA .....(((((((...--------------((......)).-(((.((.((((((((.((((((((....))).)))))..)))))))).))...)))..))))))) ( -22.70) >consensus CGAUUUUCUUGAAAAACAA_________AACCACCCAGCCAGAUCGCCAAUAAGCAGUGAAAGUGUCAACACAUUUCGAGUGCUUAUU__GCAAAUCAAACAAGAA .....((((((..........................((......)).((((((((.((((((((....))).)))))..))))))))............)))))) (-18.74 = -18.60 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:56 2006