
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,691,506 – 6,691,611 |
| Length | 105 |
| Max. P | 0.535363 |

| Location | 6,691,506 – 6,691,611 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -40.34 |
| Consensus MFE | -17.21 |
| Energy contribution | -18.93 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6691506 105 + 23771897 AUAUUC---CUCCAAGGGCA------GGUUCUCCAGAUUGGCAUGCUCGUCCUGGAGCUCCACAUCUUGCUCGGAGGGAAUGUGCU---CCUGAGCCUGCACAAUGCAAUCUUCCAG ...(((---((((..(((((------((..((((((((.((....)).)).))))))..))......)))))))))))).(((((.---.........))))).............. ( -34.00) >DroVir_CAF1 147290 108 + 1 AUAUUCCUCUUCCGCGGGCA------G---CUCCAAGCUGGGCUGCGCGUCCUGCAGCUCGACAUUUUGCUCGGCGGGCACAUGCUCGCCCUUUGCCUGCUGAAUGCACUCCUCAAG .........(((.(((((((------(---(.......(((((((((.....))))))))).......))..(((((((....)))))))...))))))).)))............. ( -42.04) >DroGri_CAF1 60775 108 + 1 AUAUUCAUCCUCAACGGGCA------G---CUCCAAGCUGGGCUGCUCGUCCUGCAGCUCGACUUUUUGCUCGGCGGGCACAUGGUCGCCCUUUGCCUGCUGAAUGCACUCCUCAAG .............(((((((------(---(((......)))))))))))..((((((..........))((((((((((...((....))..)))))))))).))))......... ( -40.50) >DroMoj_CAF1 77437 108 + 1 AUAUUCAUCCUCCGGCGGCA------U---CUCCAAGUUGGGCUGCUCAUCCUGCAGCUCAACAUUUUGCUCGGCGGGCACAUGCUCGCCCUUUGCCUGCUGAAUGCACUCCUCAAG ............((((((((------.---......((((((((((.......)))))))))).........(((((((....)))))))...)))).))))............... ( -42.60) >DroAna_CAF1 89748 108 + 1 AUAGUCGUUCUCCAUGGGCA------GGUUCUCCAGAUUGGGCUGCUCGUCCUGGAGCUCCACAUUCUGCUCGGCCGGAAUGUGCU---CCUGAGCCUGCAGAACGCAUUCCUCCAG ...(.(((((((....)(((------(((((........((((.....)))).(((((...((((((((......)))))))))))---)).))))))))))))))).......... ( -40.70) >DroPer_CAF1 66584 114 + 1 GUAGUCGUUGUCCAUGGGCAGCUCGAGGUUCUCGAGAUUGGGCUGCUCGUCCUGCAGCUCCACAUUCUGCUCGGCUGGGAUGUGCU---CCUGGGCCUGCUGAAUGCAUUCCUCCAG ..((((((((((....))))))(((((...)))))))))(((((((.......))))).)).(((((.((..(.(..(((.....)---))..).)..)).)))))........... ( -42.20) >consensus AUAUUCAUCCUCCACGGGCA______G___CUCCAAAUUGGGCUGCUCGUCCUGCAGCUCCACAUUUUGCUCGGCGGGCACAUGCU___CCUGUGCCUGCUGAAUGCACUCCUCAAG ...............(((.....................(((((((.......))))))).......(((((((((((.................))))))))..))).)))..... (-17.21 = -18.93 + 1.72)

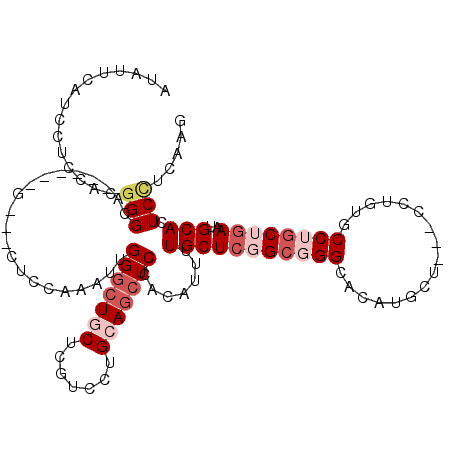

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:54 2006