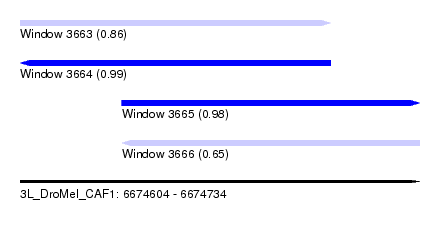
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,674,604 – 6,674,734 |
| Length | 130 |
| Max. P | 0.993431 |

| Location | 6,674,604 – 6,674,705 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6674604 101 + 23771897 CGGCGGACACACUCGAAUGCCUUAUAUCUAAAGCUUGUUUUGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUAU----AU .(((((.(......)..(((((.((((.((....(((...(((((((.((.......)).)))))))...))))).)))))))))))))).........----.. ( -22.70) >DroEre_CAF1 63310 105 + 1 CGGCGGACACACUCGAAUGCCUUAUAUCUAAAGCUUGUUUUGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUACAAGUAU .((((..(......)..))))...........((((((..(((((((.((.......)).))))))).........(((((((.....)))))))..)))))).. ( -25.80) >DroYak_CAF1 60285 101 + 1 CGGCGGACACACUCGAAUGCCUUAUAUCUAAAGCUUGUUUCGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUA----UAU .((((((((((((((((.((.(((....))).))....)))))))...))))...............((((.((....)).)))))))))........----... ( -24.10) >DroAna_CAF1 58067 96 + 1 CGGCGGACACACUCGAAUGCUUCAUAUCUAAAGCUUGUUUCGGGUUUUGUGUUUCAAAUUAUACCCAUACGA----AUGUAGGUAU-ACCUAUAUAUAU----GU ....(((((((((((((.((((........))))....)))))))...))))))...........((((...----(((((((...-.))))))).)))----). ( -22.20) >consensus CGGCGGACACACUCGAAUGCCUUAUAUCUAAAGCUUGUUUCGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUAU____AU .((((((((((((((((.((.(((....))).))....)))))))...))))..........(((.(((........))).))).)))))............... (-19.14 = -19.45 + 0.31)

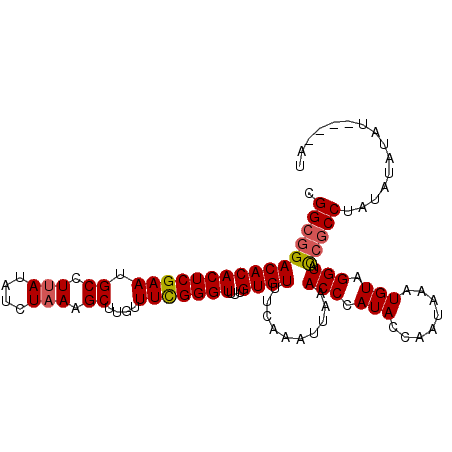
| Location | 6,674,604 – 6,674,705 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.65 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -24.48 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6674604 101 - 23771897 AU----AUAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCAAAACAAGCUUUAGAUAUAAGGCAUUCGAGUGUGUCCGCCG ..----.........(((((....(((((((.(((...(((((((.............)))))))...)))((((((....))))))....))))))).))))). ( -29.62) >DroEre_CAF1 63310 105 - 1 AUACUUGUAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCAAAACAAGCUUUAGAUAUAAGGCAUUCGAGUGUGUCCGCCG ...............(((((....(((((((.(((...(((((((.............)))))))...)))((((((....))))))....))))))).))))). ( -29.62) >DroYak_CAF1 60285 101 - 1 AUA----UAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCGAAACAAGCUUUAGAUAUAAGGCAUUCGAGUGUGUCCGCCG ...----........(((((....(((((((.(((...(((((((.............)))))))...)))((((((....))))))....))))))).))))). ( -28.92) >DroAna_CAF1 58067 96 - 1 AC----AUAUAUAUAGGU-AUACCUACAU----UCGUAUGGGUAUAAUUUGAAACACAAAACCCGAAACAAGCUUUAGAUAUGAAGCAUUCGAGUGUGUCCGCCG ..----.........(((-..((..((((----(((..(((((....((((.....)))))))))......((((((....))))))...)))))))))..))). ( -23.00) >consensus AU____AUAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCAAAACAAGCUUUAGAUAUAAGGCAUUCGAGUGUGUCCGCCG ...............(((((....(((((((.......(((((((.............)))))))......((((((....))))))....))))))).))))). (-24.48 = -24.92 + 0.44)

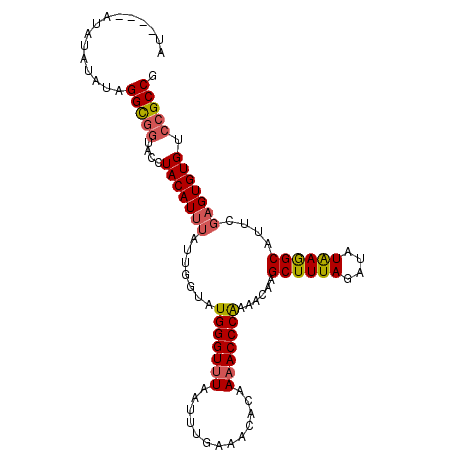
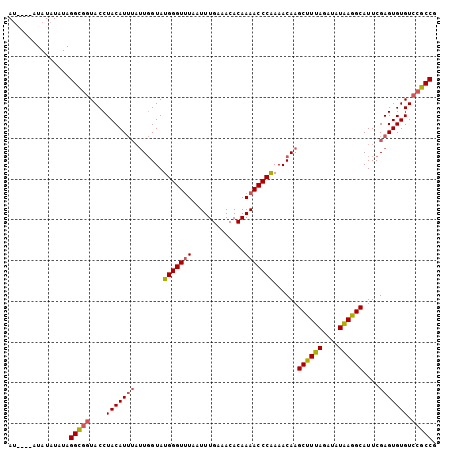
| Location | 6,674,637 – 6,674,734 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6674637 97 + 23771897 CUUGUUUUGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUAU----AUACAUAUACAUAUUU-AGUUUAUAUGCCUUU ..((...(((((((.((.......)).)))))))...))(((.(((((((.....))))))).)))----...((((((.((....-.)).))))))..... ( -18.90) >DroEre_CAF1 63343 93 + 1 CUUGUUUUGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUACAAGUAUGU-------UAUAU-AGUUUAUAUGC-UUC .(((...(((((((.((.......)).)))))))...)))...(((((((.....)))))))....((((((((-------.....-.....))))))-)). ( -20.60) >DroYak_CAF1 60318 84 + 1 CUUGUUUCGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUA----UAUAC-------UAUA------UAUAUAG-UUU ........((((((.((.......)).)))))).((((.((....)).))))....(((((((((----((...-------))))------)))))))-... ( -17.80) >DroAna_CAF1 58100 93 + 1 CUUGUUUCGGGUUUUGUGUUUCAAAUUAUACCCAUACGA----AUGUAGGUAU-ACCUAUAUAUAU----GUUCUUUUAUGUGUCCCUAUUGAUGUGCCUUU .((((...((((((((.....))))....))))..))))----....((((((-(.(...((((((----(......))))))).......).))))))).. ( -15.10) >consensus CUUGUUUCGGGUUUUGUGUUUCAAAUUAAACCCAUACCAAUAAAUGUAGGUACCGCCUAUAUAUAU____AUAC_______UAUAU_AGUUUAUAUGC_UUU ........((((((.((.......)).))))))..........(((((((.....)))))))........................................ (-14.10 = -14.35 + 0.25)

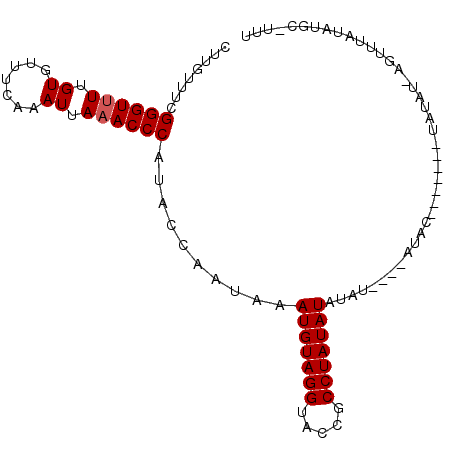
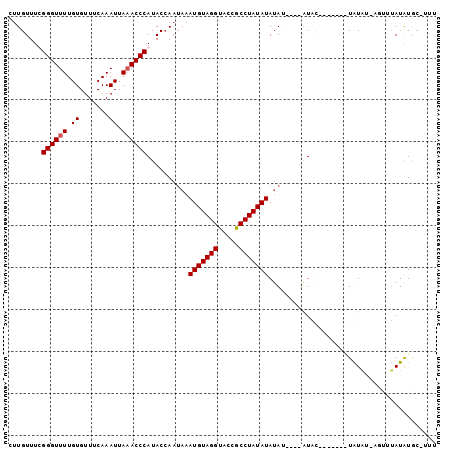
| Location | 6,674,637 – 6,674,734 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -16.77 |
| Consensus MFE | -8.92 |
| Energy contribution | -8.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6674637 97 - 23771897 AAAGGCAUAUAAACU-AAAUAUGUAUAUGUAU----AUAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCAAAACAAG ..(((..(((...((-(.((((((((....))----)))))).)))...)))))).......(((...(((((((.............)))))))...))). ( -18.12) >DroEre_CAF1 63343 93 - 1 GAA-GCAUAUAAACU-AUAUA-------ACAUACUUGUAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCAAAACAAG ...-.........((-(((((-------(((....))).)))))))...(((((..........)))))((((((.............))))))........ ( -17.22) >DroYak_CAF1 60318 84 - 1 AAA-CUAUAUA------UAUA-------GUAUA----UAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCGAAACAAG ...-(((((((------((((-------...))----)))))))))...(((((..........)))))((((((.............))))))........ ( -18.92) >DroAna_CAF1 58100 93 - 1 AAAGGCACAUCAAUAGGGACACAUAAAAGAAC----AUAUAUAUAGGU-AUACCUACAU----UCGUAUGGGUAUAAUUUGAAACACAAAACCCGAAACAAG ...............(((..............----...........(-(((((((...----.....)))))))).((((.....)))).)))........ ( -12.80) >consensus AAA_GCAUAUAAACU_AUAUA_______GUAU____AUAUAUAUAGGCGGUACCUACAUUUAUUGGUAUGGGUUUAAUUUGAAACACAAAACCCAAAACAAG ...........................................((((.....))))............(((((((.............)))))))....... ( -8.92 = -8.92 + -0.00)

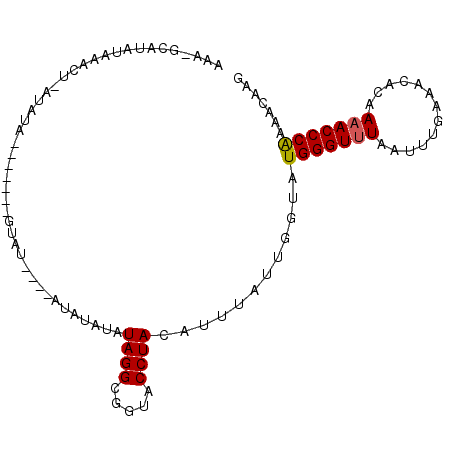
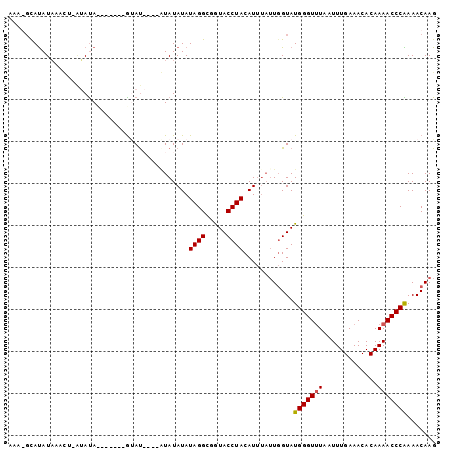
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:48 2006