
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,671,111 – 6,671,220 |
| Length | 109 |
| Max. P | 0.748818 |

| Location | 6,671,111 – 6,671,220 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.52 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -15.10 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6671111 109 + 23771897 CUUAAAAACGCAUUUCCAUUAA---CAGAAUAACAAGGCUUCCCUGAGGAGCCAUUCGGCUAUUCGGUCACGCCCC-CAACAGGGUGUUACACGCAAUCUAAACUAUACAAUC .........((...........---..(((((.(..((((((.....)))))).....).))))).((.((((((.-.....)))))).))..)).................. ( -24.20) >DroSec_CAF1 47835 109 + 1 CUUAAAAACGCAUUUUCAUUAA---CGGAAUAACAAGGCUCCCCUGAGGAGCCAUUUGGCUACUCGGUCACGCCCC-CAACCGGGUGAUACACGCAAUCUAAACUAUACAAUC .........((...........---...........((((((.....))))))...(((((....)))))(((((.-.....)))))......)).................. ( -24.90) >DroSim_CAF1 66085 109 + 1 CUUAAAAACGCAUUUUCAUUAA---CGGAAUAACAAGGCUCCCCUGAGGAGCCAUUUGGCUACUCGGUCACGCCCC-CAACAGGGUGAUACACGCAAUCUAAACUAUACAAUC .........((...........---...........((((((.....))))))...(((((....)))))(((((.-.....)))))......)).................. ( -25.00) >DroEre_CAF1 59829 109 + 1 CUUAAAAACGCAUUUCCAUUAA---CGGAACAACAAGGCUCCCCCGAGGAGCCAUUUGGAUACUUGGCCACGCCCC-UAACAAUGUGAUACACGCAAUCUAGACCAUACAUUC .............((((.....---.))))......((((((.....)))))).(((((((....(((...)))..-......((((.....))))))))))).......... ( -23.30) >DroYak_CAF1 56671 110 + 1 CUUAAAAACGCAUUUCCAUUAA---CGGAAUAACAAGACUCCCCUGAGGAGCCAUUUGGAUACUCGGCCACGCCCCUUAACAGAGUGAUACACGCAAUCUAGACUAUACAUUC .........((..((((.....---.)))).......((((...(((((.((....(((........))).)).)))))...)))).......)).................. ( -18.30) >DroAna_CAF1 54245 87 + 1 CUCAAAAACGCAUUCACACUAACAACAGCAUUACACGGCUCCCUUGAGGAC-----------CUCAGACACGCCCC-AAUCGGGGUGGCACACGCUACC-------------- ..........................(((.......((.(((.....))))-----------).......((((((-....))))))......)))...-------------- ( -19.20) >consensus CUUAAAAACGCAUUUCCAUUAA___CGGAAUAACAAGGCUCCCCUGAGGAGCCAUUUGGCUACUCGGUCACGCCCC_CAACAGGGUGAUACACGCAAUCUAAACUAUACAAUC .........((.........................((((((.....)))))).................(((((.......)))))......)).................. (-15.10 = -16.13 + 1.03)

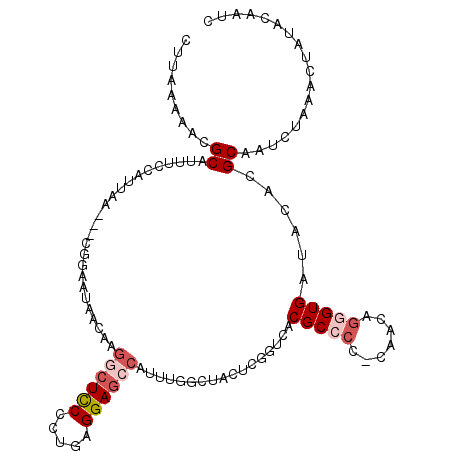

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:45 2006