
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,658,479 – 6,658,575 |
| Length | 96 |
| Max. P | 0.767868 |

| Location | 6,658,479 – 6,658,575 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
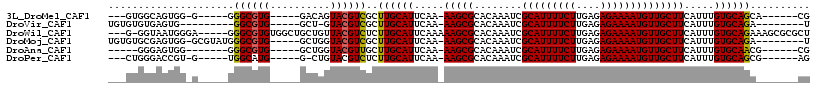
>3L_DroMel_CAF1 6658479 96 + 23771897 CG------UGCUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUU-UUGAAUGCAAGCGACGUACUGUC-----CACGCCC-----C-CCACUGCCAC--- .(------(((.((((((((...(((...((((((....))))))))).))))))))((((-........))))...))))....-----.......-----.-..........--- ( -22.20) >DroVir_CAF1 102031 93 + 1 A--------UCUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUU-UUGAAUGCAAGCGACGUAC-AGC-----CACGCC---------CACUCACACACA .--------...((((((((...(((...((((((....))))))))).))))))))((((-........))))..(((..-...-----.)))..---------............ ( -18.50) >DroWil_CAF1 43003 108 + 1 AGCGCGCUUUCUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUUUUUGAAUGCAAGAGACGUAACAGCAGCCACACGCCC-----UCCCAUUACC-C--- .(((.(....((((((((((...(((...((((((....))))))))).))))))(((..(((((....)))))..)))....))))...).)))..-----..........-.--- ( -23.40) >DroMoj_CAF1 39684 102 + 1 A--------UCUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUU-UUGAAUGCAAGCGACGUACCAGC-----CACGCCCAUACGC-CCACUCGCACACA .--------...((((((((...(((...((((((....))))))))).))))))))((..-......))..((((((((...((-----...))...)))).-....))))..... ( -23.50) >DroAna_CAF1 40378 93 + 1 CG------CGUUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUU-UUGAAUGCAAGCAACGUACCAGC-----CACGCCC-------CCACUCCC----- .(------((((((((((((...(((...((((((....))))))))).))))))))((((-........)))))))))......-----.......-------........----- ( -22.50) >DroPer_CAF1 30126 95 + 1 CU------CGCUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUU-UUGAAUGCAAGAGACGUACAG-C-----CAUGCCA-----C-ACGGUCCCAG--- .(------((((((.........)))..(((((((....))))))))))).((((((((((-(((....)))))).)))))))-.-----...(((.-----.-..))).....--- ( -24.20) >consensus AG_______GCUGCACAAAUGAAGCAACAUUUUCUCUCAAGAAAAUGCGAUUUGUGCGCUU_UUGAAUGCAAGCGACGUACCAGC_____CACGCCC_____C_CCACUCCCAC___ ............((((((((...(((...((((((....))))))))).))))))))((.........))............................................... (-16.20 = -16.20 + -0.00)



| Location | 6,658,479 – 6,658,575 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -20.99 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6658479 96 - 23771897 ---GUGGCAGUGG-G-----GGGCGUG-----GACAGUACGUCGCUUGCAUUCAA-AAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAGCA------CG ---(((.(.(..(-(-----.((((((-----.....)))))).))..)......-....((((((((.(((((((((....)))))))))......)))))))).)))------). ( -33.20) >DroVir_CAF1 102031 93 - 1 UGUGUGUGAGUG---------GGCGUG-----GCU-GUACGUCGCUUGCAUUCAA-AAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAGA--------U ((.(((..((((---------(.((((-----...-.)))))))))..))).)).-....((((((((.(((((((((....)))))))))......))))))))...--------. ( -31.60) >DroWil_CAF1 43003 108 - 1 ---G-GGUAAUGGGA-----GGGCGUGUGGCUGCUGUUACGUCUCUUGCAUUCAAAAAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAGAAAGCGCGCU ---.-.(.((((.((-----(((((((..((....)))))))).))).)))))....(((((((((((.(((((((((....)))))))))......))))))((.....))))))) ( -31.90) >DroMoj_CAF1 39684 102 - 1 UGUGUGCGAGUGG-GCGUAUGGGCGUG-----GCUGGUACGUCGCUUGCAUUCAA-AAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAGA--------U ((.((((((((((-.(((((.(((...-----))).))))))))))))))).)).-....((((((((.(((((((((....)))))))))......))))))))...--------. ( -41.00) >DroAna_CAF1 40378 93 - 1 -----GGGAGUGG-------GGGCGUG-----GCUGGUACGUUGCUUGCAUUCAA-AAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAACG------CG -----..(((((.-------(((((..-----((......))))))).)))))..-..((((((((((.(((((((((....)))))))))......))))))))...)------). ( -27.80) >DroPer_CAF1 30126 95 - 1 ---CUGGGACCGU-G-----UGGCAUG-----G-CUGUACGUCUCUUGCAUUCAA-AAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAGCG------AG ---..(((((.((-(-----..((...-----)-)..))))))))..........-..((((((((((.(((((((((....)))))))))......)))))))).)).------.. ( -30.30) >consensus ___GUGGGAGUGG_G_____GGGCGUG_____GCUGGUACGUCGCUUGCAUUCAA_AAGCGCACAAAUCGCAUUUUCUUGAGAGAAAAUGUUGCUUCAUUUGUGCAGA_______CG .....................((((((..........))))))..((((((.....(((((........(((((((((....)))))))))))))).....)))))).......... (-20.99 = -20.88 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:42 2006