
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,641,870 – 6,642,042 |
| Length | 172 |
| Max. P | 0.981450 |

| Location | 6,641,870 – 6,641,964 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -28.19 |
| Energy contribution | -28.55 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6641870 94 + 23771897 GCAUUAAUAAGCGGUCAAAGUUUUGAUGGCUUAAUCGAUAU---GGCGACGUCGGCAAC---UUCUGCUCGGCGACCGCAGCGCAUACUGCGGUCAC-CGC (((....(((((.(((((....))))).)))))...((..(---(.((....)).))..---.))))).(((.((((((((......)))))))).)-)). ( -37.30) >DroSec_CAF1 18987 94 + 1 GCAUUAAUAAGCGGUCAAAGUUUUGAUGGCUUAAUCGAUAC---GGCGACGUCGGCAAC---UUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCAC-CGC .......(((((.(((((....))))).))))).......(---((....(((((((..---...))).))))((((((((......)))))))).)-)). ( -37.00) >DroSim_CAF1 34973 94 + 1 GCAUUAAUAAGCGGUCAAAGUUUUGAUGGCUUAAUCGAUAC---GGCGACGUCGGCAAC---UUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCAC-CGC .......(((((.(((((....))))).))))).......(---((....(((((((..---...))).))))((((((((......)))))))).)-)). ( -37.00) >DroEre_CAF1 28897 87 + 1 GCAUUAAUAAGCGGUCAAAGUUUUGAUGGCUUAAUCGAUAU---GACGACGUCGGCAAC---UUCUGCUCGGCGACCACAGCGCAUUCUGCGG-------- ((.....(((((.(((((....))))).))))).(((....---..)))((((((((..---...))).)))))......))((.....))..-------- ( -25.50) >DroYak_CAF1 26564 94 + 1 GCAUUAAUAAGCGGUCAAAGUUUUGAUGGCUUAAUCGAUAU---GACGACGUCGGCAAC---UUCUGCUCGGCGACCACAGCGCAUUCUGCGGUCAC-CGC (((....(((((.(((((....))))).))))).(((((..---......)))))....---...))).(((.((((.(((......))).)))).)-)). ( -30.00) >DroAna_CAF1 22037 101 + 1 ACAUUAAUAAGCGGUCGAAGUUUUGAUCGUUUAAUCGAUAGGAUGGAGACUCCCGCACUGGAAUUUGUCUGGCGACCGCAGAGCAUUCUGUGGUCGCCCGC .......(((((((((((....))))))))))).......(((..((...(((......))).))..)))(((((((((((......)))))))))))... ( -40.50) >consensus GCAUUAAUAAGCGGUCAAAGUUUUGAUGGCUUAAUCGAUAU___GGCGACGUCGGCAAC___UUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCAC_CGC .......(((((.(((((....))))).))))).................(((((((........))).))))((((((((......))))))))...... (-28.19 = -28.55 + 0.36)


| Location | 6,641,926 – 6,642,042 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Mean single sequence MFE | -41.53 |
| Consensus MFE | -20.65 |
| Energy contribution | -21.90 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6641926 116 + 23771897 -UUCUGCUCGGCGACCGCAGCGCAUACUGCGGUCAC-CGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGCCGAGCAA-GGAUUAGGUAAUCAGCAC-UUUAAUUGCGAGUAAA -...(((((((.((((((((......)))))))).)-)((((.((((......((.(((....))).)).....))))))))..-.((((....)))).(((.-......)))))))).. ( -42.90) >DroSec_CAF1 19043 116 + 1 -UUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCAC-CGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGCCGAGCAA-GGAUUAGGUAAUCAGCAC-UUUAAUUGCGAGUAAA -...(((((((.((((((((......)))))))).)-)((((.((((......((.(((....))).)).....))))))))..-.((((....)))).(((.-......)))))))).. ( -42.30) >DroSim_CAF1 35029 116 + 1 -UUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCAC-CGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGCCGAGCAA-GGAUUAGGUAAUCAGCAC-UUUAAUUGCGAGUAAA -...(((((((.((((((((......)))))))).)-)((((.((((......((.(((....))).)).....))))))))..-.((((....)))).(((.-......)))))))).. ( -42.30) >DroEre_CAF1 28953 97 + 1 -UUCUGCUCGGCGACCACAGCGCAUUCUGCGG----------------UGAAAAGUAUGCCAGCAUUCUGGUCAGGACGAGCAA-GGAUUAGGGAAUCAGCAC-UUUAAUUGCAAC---- -...((((((..((((.((.(((.....))).----------------))...((.(((....))).))))))....)))))).-.((((((((........)-))))))).....---- ( -28.00) >DroYak_CAF1 26620 112 + 1 -UUCUGCUCGGCGACCACAGCGCAUUCUGCGGUCAC-CGCUCCGGCGCUGAAAAGUAUGCCAGCAUUCUGGUCACGACAAGCAA-GGAUUAGGUAAUCAGCGC-UUUAAUUGCCAG---- -...((((.((((((..((((((.....((((...)-)))....))))))....)).))))))))..(((((......((((..-.((((....))))...))-)).....)))))---- ( -37.00) >DroAna_CAF1 22098 99 + 1 AAUUUGUCUGGCGACCGCAGAGCAUUCUGUGGUCGCCCGCUGCCUGGCUCGCCAGUCUGAU-----------CCGGAGACGGACUGGCUUAGGC-GGCAGGACCUCCAAUU--------- .....(((((((((((((((......))))))))))).(((((((((...(((((((((.(-----------(....)))))))))))))))))-))).))))........--------- ( -56.70) >consensus _UUCUGCUCGGCGACCGCAGCGCAUUCUGCGGUCAC_CGCUCCGGCGCUCAAAAGUAUGCUAGCAUUCUGGUCACGACGAGCAA_GGAUUAGGUAAUCAGCAC_UUUAAUUGCGAG____ ....((((((.(((((((((......))))))))........................(((((....)))))...).))))))...((((....)))).(((........)))....... (-20.65 = -21.90 + 1.25)



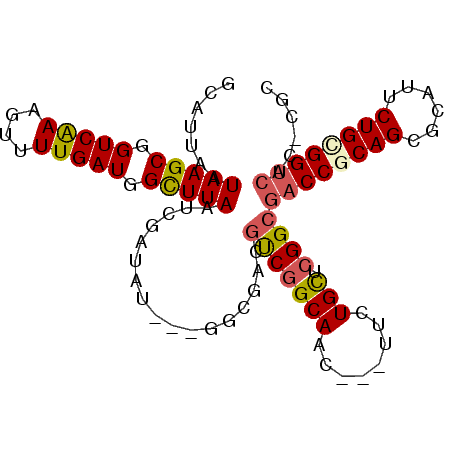
| Location | 6,641,926 – 6,642,042 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.40 |
| Mean single sequence MFE | -41.98 |
| Consensus MFE | -23.39 |
| Energy contribution | -26.03 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6641926 116 - 23771897 UUUACUCGCAAUUAAA-GUGCUGAUUACCUAAUCC-UUGCUCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCG-GUGACCGCAGUAUGCGCUGCGGUCGCCGAGCAGAA- ................-.((((.............-..(((((((((....((.(((....))).)).....))))).))))(-(((((((((((....)))))))))))).))))...- ( -46.00) >DroSec_CAF1 19043 116 - 1 UUUACUCGCAAUUAAA-GUGCUGAUUACCUAAUCC-UUGCUCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCG-GUGACCGCAGAAUGCGCUGCGGUCGCCGAGCAGAA- ................-.((((.............-..(((((((((....((.(((....))).)).....))))).))))(-((((((((((......))))))))))).))))...- ( -44.70) >DroSim_CAF1 35029 116 - 1 UUUACUCGCAAUUAAA-GUGCUGAUUACCUAAUCC-UUGCUCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCG-GUGACCGCAGAAUGCGCUGCGGUCGCCGAGCAGAA- ................-.((((.............-..(((((((((....((.(((....))).)).....))))).))))(-((((((((((......))))))))))).))))...- ( -44.70) >DroEre_CAF1 28953 97 - 1 ----GUUGCAAUUAAA-GUGCUGAUUCCCUAAUCC-UUGCUCGUCCUGACCAGAAUGCUGGCAUACUUUUCA----------------CCGCAGAAUGCGCUGUGGUCGCCGAGCAGAA- ----...(((......-.)))..............-(((((((...((.((((....))))))........(----------------((((((......)))))))...)))))))..- ( -25.30) >DroYak_CAF1 26620 112 - 1 ----CUGGCAAUUAAA-GCGCUGAUUACCUAAUCC-UUGCUUGUCGUGACCAGAAUGCUGGCAUACUUUUCAGCGCCGGAGCG-GUGACCGCAGAAUGCGCUGUGGUCGCCGAGCAGAA- ----((((((....((-(((..((((....)))).-.)))))....)).))))..(((((((............)))....((-((((((((((......))))))))))))))))...- ( -44.30) >DroAna_CAF1 22098 99 - 1 ---------AAUUGGAGGUCCUGCC-GCCUAAGCCAGUCCGUCUCCGG-----------AUCAGACUGGCGAGCCAGGCAGCGGGCGACCACAGAAUGCUCUGCGGUCGCCAGACAAAUU ---------........((((((((-......(((((((.(((....)-----------))..)))))))......)))))..(((((((.((((....)))).))))))).)))..... ( -46.90) >consensus ____CUCGCAAUUAAA_GUGCUGAUUACCUAAUCC_UUGCUCGGCGUGACCAGAAUGCUAGCAUACUUUUGAGCGCCGGAGCG_GUGACCGCAGAAUGCGCUGCGGUCGCCGAGCAGAA_ .....................................((((((((((....((.(((....))).)).....))))).......((((((((((......)))))))))).))))).... (-23.39 = -26.03 + 2.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:39 2006