
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,637,893 – 6,638,005 |
| Length | 112 |
| Max. P | 0.971866 |

| Location | 6,637,893 – 6,637,984 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.85 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6637893 91 - 23771897 ACUGCAAAAGAUAAACGUCCACUUGGUUCGCUUGGCUGGCCGAGCGAAAAACAAAAGACC-GA--AAAGGGUCAACAAUGGCCACG----GCGGAUUG- .((((...........(((...(((.(((((((((....)))))))))...)))..))).-..--....(((((....)))))...----))))....- ( -29.90) >DroVir_CAF1 74113 76 - 1 ACUGCAAAAGAUAUAUGCCCACUUGGC--GCUUGGCUGGCCGAGCGCAAAACUCAAGACA-----AAAGGGUCAACAAUGGAG---------------- ...(((.........)))(((.(((((--((((((....)))))))).........(((.-----.....)))..))))))..---------------- ( -22.80) >DroPse_CAF1 6136 86 - 1 ACUGCAAAAGAUAAACGUCCACUUGGCGCGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA--AAAGGGUCAACAAUGG----------AGGAUUG- ...............(.((((.(((...(((((((....)))))))..........(((((..--...)))))..))))))----------).)....- ( -28.40) >DroGri_CAF1 6710 97 - 1 ACUGCAAAAGAUAUAUGUCCACUUGGC--GCUUGGCUGGUCGAGCGCAAAGCUCAAGACACAAAAAAAGGGUCAACAAUGGAGGCGAGCAGUGAAUUCC (((((..........((((..(((.((--((((((....)))))))).))).....))))..........(((.........)))..)))))....... ( -24.70) >DroYak_CAF1 22420 92 - 1 ACUGCAAAAGAUAAACGUCCACUUGGCUCGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA--AAAGGGUCAACAAUGGCCACG----UUGGAUUG- ................((((((.((((((((((((....)))))))).........(((((..--...))))).......)))).)----.)))))..- ( -33.80) >DroPer_CAF1 6206 86 - 1 ACUGCAAAAGAUAAACGUCCACUUGGCGCGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA--AAAGGGUCAACAAUGG----------AGGAUUG- ...............(.((((.(((...(((((((....)))))))..........(((((..--...)))))..))))))----------).)....- ( -28.40) >consensus ACUGCAAAAGAUAAACGUCCACUUGGC_CGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA__AAAGGGUCAACAAUGG__________AGGAUUG_ ..................(((.(((...(((((((....)))))))..........(((((.......)))))..)))))).................. (-17.99 = -18.85 + 0.86)

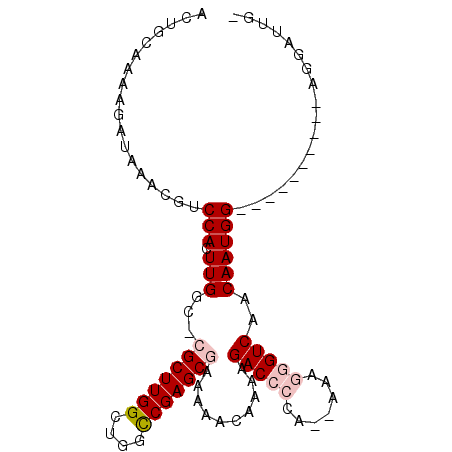
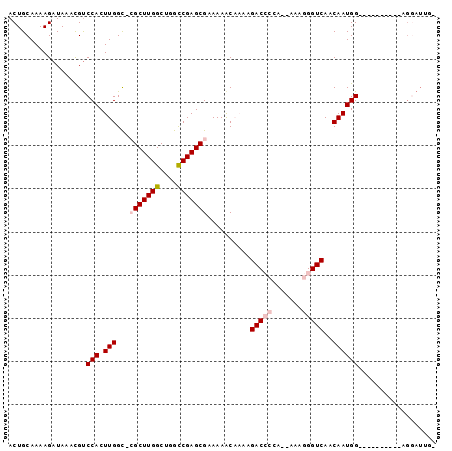
| Location | 6,637,913 – 6,638,005 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -16.02 |
| Energy contribution | -16.91 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6637913 92 - 23771897 GCCA---------------GCUAGAAGU--GCACAGUGACUGCAAAAGAUAAACGUCCACUUGGUUCGCUUGGCUGGCCGAGCGAAAAACAAAAGACC-GA--AAAGGGUCA ((((---------------((.......--))..((((((..............)).))))))))((((((((....)))))))).........((((-..--....)))). ( -26.04) >DroVir_CAF1 74122 78 - 1 ---------------------------CACAUACAGUGACUGCAAAAGAUAUAUGCCCACUUGGC--GCUUGGCUGGCCGAGCGCAAAACUCAAGACA-----AAAGGGUCA ---------------------------(((.....)))................((((..(((((--((((((....))))))))....(....).))-----)..)))).. ( -22.00) >DroPse_CAF1 6150 108 - 1 GCAAACUAACUAAAAUUUCGCUGCAAGU--GCACAGUGACUGCAAAAGAUAAACGUCCACUUGGCGCGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA--AAAGGGUCA ...............((((((..(((((--(.((.((..((.....))....)))).))))))..))((((((....)))))))))).......(((((..--...))))). ( -31.30) >DroGri_CAF1 6735 88 - 1 GC----------------------AAGUACACACAGUGACUGCAAAAGAUAUAUGUCCACUUGGC--GCUUGGCUGGUCGAGCGCAAAGCUCAAGACACAAAAAAAGGGUCA ((----------------------(.((.(((...))))))))..........((((..(((.((--((((((....)))))))).))).....)))).............. ( -21.50) >DroYak_CAF1 22440 93 - 1 GCCA---------------GCUAGAAGU--GCACAGUGACUGCAAAAGAUAAACGUCCACUUGGCUCGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA--AAAGGGUCA ((((---------------((.......--))..((((((..............)).))))))))((((((((....)))))))).........(((((..--...))))). ( -30.94) >DroPer_CAF1 6220 108 - 1 GCAAACUAACUAAAAUUUCGCUGCAAGU--GCACAGUGACUGCAAAAGAUAAACGUCCACUUGGCGCGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA--AAAGGGUCA ...............((((((..(((((--(.((.((..((.....))....)))).))))))..))((((((....)))))))))).......(((((..--...))))). ( -31.30) >consensus GC_A_______________GCU__AAGU__GCACAGUGACUGCAAAAGAUAAACGUCCACUUGGC_CGCUUGGCUGGCCGAGCGAAAAACAAAAGACCCCA__AAAGGGUCA ...............................((.((((((..............)).))))))...(((((((....)))))))..........(((((.......))))). (-16.02 = -16.91 + 0.89)

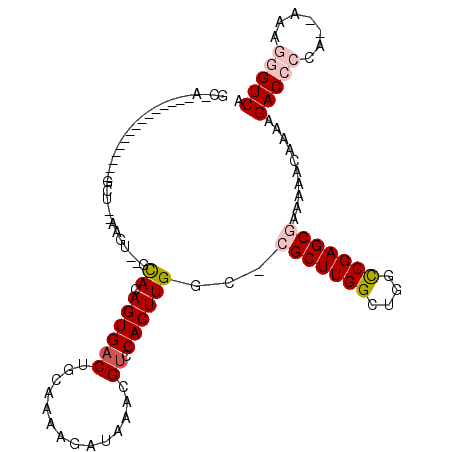
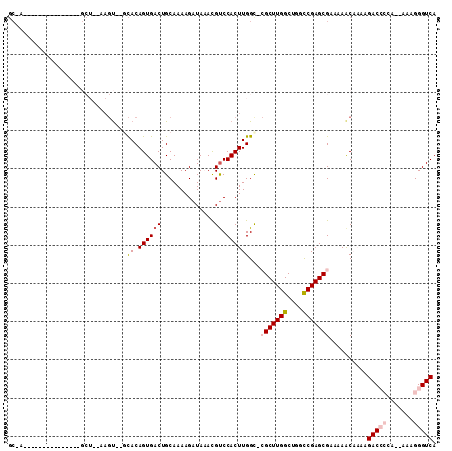
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:36 2006