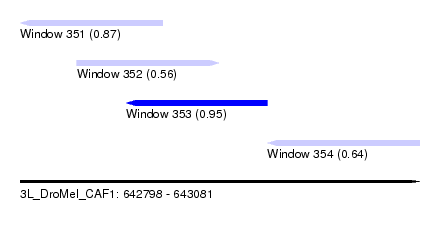
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 642,798 – 643,081 |
| Length | 283 |
| Max. P | 0.945580 |

| Location | 642,798 – 642,899 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.97 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -12.74 |
| Energy contribution | -15.30 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 642798 101 - 23771897 UCGGCA--------------GCCAACGAGUCUAUCAAGGCUUGACAACAUUUGCAUCUGGUCCG----GGCCAACUUG-AGAUAACUCGACCACAUCAUGGGGCAAGUUGCAGCGGCUGC ...(((--------------(((..(((((((....))))))).......(((((.((.((((.----(......(((-((....)))))........).)))).)).))))).)))))) ( -34.34) >DroSec_CAF1 9950 101 - 1 UCGGCA--------------GCCAACGAGUCUAUCAAGACUUGACAACAUUUGCAUCUGGUCCG----GGCCAACUUG-AGAUAACUCGACCAGAUCAUGGGGCAAGUUGCAGCGGCUGC ...(((--------------(((..(((((((....))))))).((((..((((((((((((.(----(.....)).(-((....)))))))))))......))))))))....)))))) ( -36.60) >DroEre_CAF1 12968 101 - 1 UCGGCA--------------GCCAACAAGUCUAUCAAGGCAUGACAACAUUUGCAUCUGGUCCG----GGCCAACUUG-AGAUAACUCGACCACAUCAUGUGGCAAGUUGCAGCGGCUGC ...(((--------------(((..........((((((((..........)))...((((...----.)))).))))-)(.(((((.(.((((.....))))).))))))...)))))) ( -32.30) >DroWil_CAF1 10214 109 - 1 UUGGCAACCAUUUGGUUACUGCCAACAAGUCUAUCAAGACCUGACGACAUUUGCCUCCUAUCUGUUGUAGCCAGCUUGCAGCCUGGUGGGCCACUUUAG------UGUUGUAACU----- (((((((((....)))...))))))((.((((....)))).))((((((((.....((((((.(((((((.....)))))))..)))))).......))------))))))....----- ( -33.50) >DroYak_CAF1 10252 101 - 1 UCGGCA--------------GCCAACAAGUCUAUCAAGACUUGACAACAUUUGCAUCUGGACGG----GGCCAACUCG-AGAUAACUCGACCACAUCGUGGGGCAACUUGCUGCGGCUGC ...(((--------------(((..(((((((....))))))).........(((...(.(.(.----.(((...(((-((....)))))((((...)))))))..).).)))))))))) ( -37.40) >consensus UCGGCA______________GCCAACAAGUCUAUCAAGACUUGACAACAUUUGCAUCUGGUCCG____GGCCAACUUG_AGAUAACUCGACCACAUCAUGGGGCAAGUUGCAGCGGCUGC ..(((...............)))..(((((((....))))))).((((..((((...(((((......))))).................(((.....))).)))))))).......... (-12.74 = -15.30 + 2.56)

| Location | 642,838 – 642,939 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -32.49 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 642838 101 + 23771897 UCAAGUUGGCCCGGACCAGAUGCAAAUGUUGUCAAGCCUUGAUAGACUCGUUGGCUGCCGACGCGAUUUGUCAACGACUCCACGGCGGCUGGUUUGGGUUG .......((((((((((((.(((...((..(((.....(((((((((.((((((...)))))).).)))))))).)))..))..))).)))))))))))). ( -39.60) >DroSec_CAF1 9990 101 + 1 UCAAGUUGGCCCGGACCAGAUGCAAAUGUUGUCAAGUCUUGAUAGACUCGUUGGCUGCCGACGCGAUUUGUCAACGACUCCACGGCGGCUGGUUUGGGUUG .......((((((((((((.......((((((..(((((((((((((.((((((...)))))).).)))))))).))))..)))))).)))))))))))). ( -42.80) >DroEre_CAF1 13008 101 + 1 UCAAGUUGGCCCGGACCAGAUGCAAAUGUUGUCAUGCCUUGAUAGACUUGUUGGCUGCCGACGCGAUUUGUCAACGACUCCACGGCGGCUGGUUUGAAUUG (((((..((((..(((.(.((....)).).)))..(((.((...((.(((((((((((....)))....)))))))).)))).)))))))..))))).... ( -30.10) >DroYak_CAF1 10292 101 + 1 UCGAGUUGGCCCCGUCCAGAUGCAAAUGUUGUCAAGUCUUGAUAGACUUGUUGGCUGCCGACGCGAUUUGUCAACGACUCCGCGGCGGCUGGUUUGGAUUG .............((((((((...........(((((((....)))))))..((((((((.((.((.(((....))).)))))))))))).)))))))).. ( -35.80) >consensus UCAAGUUGGCCCGGACCAGAUGCAAAUGUUGUCAAGCCUUGAUAGACUCGUUGGCUGCCGACGCGAUUUGUCAACGACUCCACGGCGGCUGGUUUGGAUUG .......((((((((((((.(((...((..(((.....(((((((((.((((((...)))))).).)))))))).)))..))..))).)))))))))))). (-32.49 = -33.30 + 0.81)


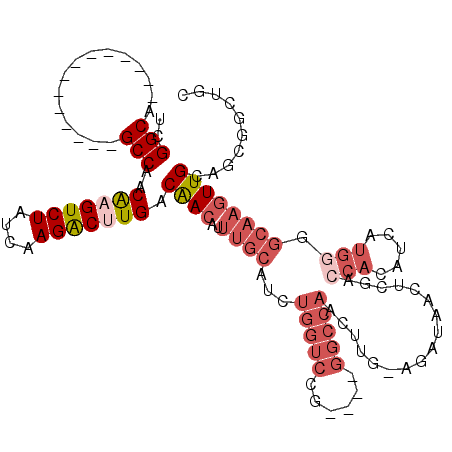
| Location | 642,873 – 642,973 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -17.15 |
| Consensus MFE | -16.37 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 642873 100 - 23771897 UGCCCUCACCACAUCACUCCACUCCAAACUU-AAGCAACCCAAACCAGCCGCCGUGGAGUCGUUGACAAAUCGCGUCGGCAGCCAACGAGUCUAUCAAGGC .((((((........(((((((.........-.....................))))))).((((((.......)))))).......)))........))) ( -18.41) >DroEre_CAF1 13043 80 - 1 UCAC---------------CACUCCAAA------CCAAUUCAAACCAGCCGCCGUGGAGUCGUUGACAAAUCGCGUCGGCAGCCAACAAGUCUAUCAAGGC ....---------------.........------.............((((.(((((.(((...)))...))))).)))).(((..............))) ( -15.44) >DroYak_CAF1 10327 101 - 1 UCACCUGACCACUUUACUCCACUCCAAACCAAAACCAAUCCAAACCAGCCGCCGCGGAGUCGUUGACAAAUCGCGUCGGCAGCCAACAAGUCUAUCAAGAC ...............................................((((.(((((.(((...)))...))))).)))).........((((....)))) ( -17.60) >consensus UCACCU_ACCAC_U_ACUCCACUCCAAAC___AACCAAUCCAAACCAGCCGCCGUGGAGUCGUUGACAAAUCGCGUCGGCAGCCAACAAGUCUAUCAAGGC ...............................................((((.(((((.(((...)))...))))).)))).........((((....)))) (-16.37 = -15.93 + -0.44)

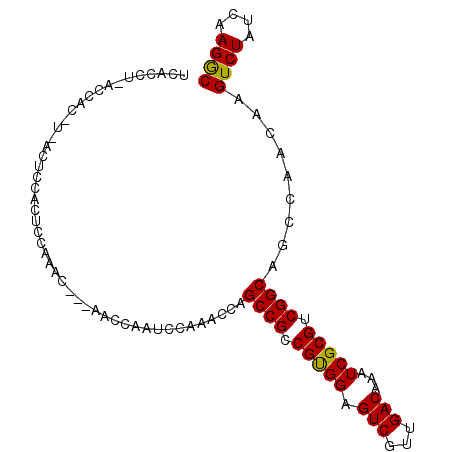
| Location | 642,973 – 643,081 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.27 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -11.98 |
| Energy contribution | -13.29 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 642973 108 - 23771897 -UGAAAUGUUUGCAAGAGGGAGUGCACUAAGCUGUAUCUGUCUAUUAUAAAACGUUCAUAG-----------GGAUAGCCCAUGCAAUGAAAACUCAGCACUCCCAAGAAUCUGGGCACC -.........(((.(((((((((((.....(((((..((((..((........))..))))-----------..)))))........(((....))))))))))).....)))..))).. ( -27.90) >DroSec_CAF1 10130 101 - 1 -UGAAAUGUUUGCAAGAGGGAGUGUACUAAACUGUAGCUAUCUCUA-------GUUCAUAG-----------GAAUAACCCAUGCAAAGAAAACUCAGCACUCCCAAGAAUCUGGGCACC -.........(((.(((((((((((......(((((((((....))-------))).))))-----------................((....)).)))))))).....)))..))).. ( -22.50) >DroEre_CAF1 13123 94 - 1 UUUAAAUGGUUGCAAUAGGGAGUGUUAUAAAAAACC---------------ACGUACAUGG-----------GAAUAGUCCAUGUAAAGACAACUCAGCACUCCCAAGAAUCUGGGCACC .......(((.((....(((((((((..........---------------...(((((((-----------(.....))))))))..((....))))))))))).((...))..))))) ( -25.20) >DroYak_CAF1 10428 108 - 1 ------------CAAUAAGCAGGGCUAUAAAGUAUGGCUGUCUAUUAUGAAAUGUACAUGGGAAUAACACGGGAAUAACCCGUGUAAAGACAACUCAGCACUCCCUACAAUCUGAGCACC ------------......((..(((((((...))))))).............((((...((((...(((((((.....)))))))...((....)).....))))))))......))... ( -25.70) >consensus _UGAAAUGUUUGCAAGAGGGAGUGCAAUAAACUAUAGCUGUCUAUU_____ACGUACAUAG___________GAAUAACCCAUGCAAAGAAAACUCAGCACUCCCAAGAAUCUGGGCACC .................((((((((.............................(((((((..................)))))))..((....)).))))))))............... (-11.98 = -13.29 + 1.31)

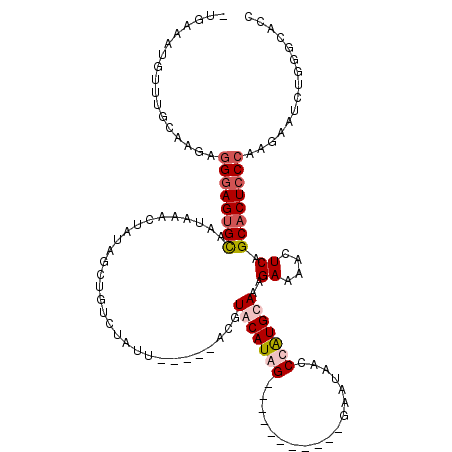
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:06 2006