| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,631,558 – 6,631,669 |
| Length | 111 |
| Max. P | 0.946004 |

| Location | 6,631,558 – 6,631,669 |
|---|---|
| Length | 111 |
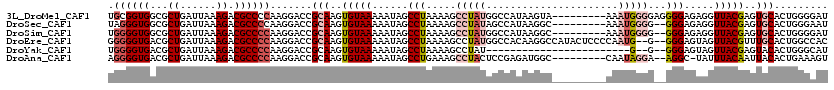
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -37.12 |
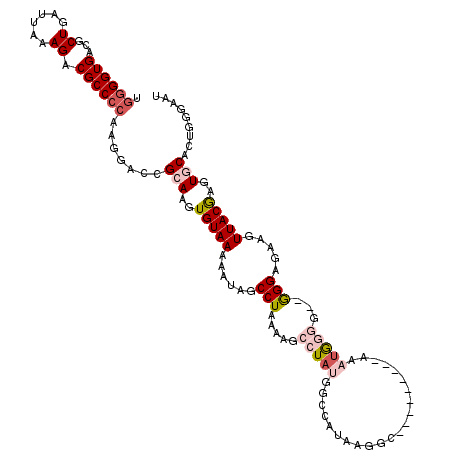
| Consensus MFE | -18.60 |
| Energy contribution | -20.02 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6631558 111 - 23771897 UGCGGUGGCGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUAAAAGCCUAUGGCCAUAAGUA---------AAAUGGGGAGGGGAGAGGUUACGAGUGCACUGGGGAU ((((((((((((......)).))))(....)))))))((((((....((((((.....(((....((((.....---------..))))....)))..))))))....))))))...... ( -36.00) >DroSec_CAF1 8575 109 - 1 UAGGGUGGCGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUAAAAGCCUAUAGCCAUAAGGC---------AAAUGGGG--GGGAGAGGUUACGAGUGCACUGGGAAU ...((.((((((......)).)))))).....((.((.(((((....((((((.....(((((.(((....)))---------..))))).--.....))))))....)))))))))... ( -35.30) >DroSim_CAF1 24505 109 - 1 UGGGGUGGCGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUAAAAGCCUAUGGCCAUAAGGC---------AAAUGGGG--GGGAGAGGUUACGAGUGCACUGGGGAU (((((((...((......)).)))))))....((.((.(((((....((((((.....(((((.(((....)))---------..))))).--.....))))))....))))))).)).. ( -40.70) >DroEre_CAF1 18498 116 - 1 GGGGGUGACGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUAAAAGCCUAUGGCCACAAGGCCAUACUCCCCAAUG--G--GGGAGUAGUUACGUUUGCACUGGCCAC .((((((...((......)).))))))..((.((...((((((((..((((...........(((((....)))))((((((((....--)--)))))))))))..)))))))))))).. ( -50.30) >DroYak_CAF1 11898 92 - 1 UGGGGUGACGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUAAAAGCCUAU------------------------G--G--GGGAGUAGUUACGAGUACACUGGGCAU (((((((...((......)).)))))))......((.((((((....((((.((....(((..------------------------.--.--)))..))))))....))))))..)).. ( -27.10) >DroAna_CAF1 10526 108 - 1 AGGGGUGACGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUGAAAGCCUACUCCGAGAUGGC---------CAAUAGGA--AGGC-UAUUUACAAUUACACUGAAAGU .((((((...((......)).))))))..........((((((((((((((((.....((((..((.....)).---------...)))).--))))-)))))....)))))))...... ( -33.30) >consensus UGGGGUGACGCUGAUUAAAGACGCCCCAAGGACCGCAAGUGUAAAAAUAGCCUAAAAGCCUAUGGCCAUAAGGC_________AAAUGGGG__GGGAGAAGUUACGAGUGCACUGGGAAU .((((((...((......)).)))))).......(((..(((((......(((.....(((((......................)))))...))).....)))))..)))......... (-18.60 = -20.02 + 1.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:34 2006