
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,624,883 – 6,625,033 |
| Length | 150 |
| Max. P | 0.962468 |

| Location | 6,624,883 – 6,624,993 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6624883 110 + 23771897 UAGGGAUAUCUC--ACAUGGGUAUGUGUGCUUAUGUGUAUGU--------GCGCCCAUUCAGUAUACUCGACAUUUGGGUCUGCUUUAAAGCGUUAAUUCGCUUUCGAGGGUGUGUAUAA ...........(--(((((((((....))))))))))((((.--------.(((((....((((.((((((...)))))).))))..((((((......))))))...)))))..)))). ( -37.20) >DroSec_CAF1 1919 110 + 1 UAGGGAUAUCUC--ACAUGGGUAUGUGUGCCUAUGUGUGUGU--------GCGCCCAUUCAGUAUACUCGACAUUUGGGUCUGCUUUAAAGCGUUAAUUCGCUUUCGAGGGUGUGUAUAA ...........(--(((((((((....))))))))))((((.--------.(((((....((((.((((((...)))))).))))..((((((......))))))...)))))..)))). ( -40.00) >DroSim_CAF1 10060 110 + 1 UAGGGAUAUCUC--ACAUGGGUAUGUGUGCCUAUGUGUGUGU--------GCGCCCAUUCAGUAUACUCGACAUUUGGGUCUGCUUUAAAGCGUUAAUUCGCUUUCGAGGGUGUGUAUAA ...........(--(((((((((....))))))))))((((.--------.(((((....((((.((((((...)))))).))))..((((((......))))))...)))))..)))). ( -40.00) >DroYak_CAF1 3955 94 + 1 UAGGGAU-UCUC--ACAUGGAUAUGUGCGCUUAUGUGUGGG-----------------------UACUCGACAUUUGGGUCUGCUUUAAAGCGUUAAUUCGGUUUCGAGGGUGUGUAUAA ((.(.((-((((--..((.((.....((((((....(..(.-----------------------.((((((...)))))))..)....))))))....)).))...)))))).).))... ( -21.40) >DroAna_CAF1 2156 117 + 1 UGGGGUGAGCACAUACUUGGGCAAGGAUACCCGUGUGUGUAUAGGAUGUGGUGGGCAUCCA-CACUCCCGACAU-UGGGUCAG-UUUAAAGCGUUAAUUCGCUUUUGAGGGUGUGUAUAA ........((((((((..(((........)))))))))))...((((((.....))))))(-(((.((((((..-...)))..-((.((((((......)))))).))))).)))).... ( -40.80) >consensus UAGGGAUAUCUC__ACAUGGGUAUGUGUGCCUAUGUGUGUGU________GCGCCCAUUCAGUAUACUCGACAUUUGGGUCUGCUUUAAAGCGUUAAUUCGCUUUCGAGGGUGUGUAUAA ..............(((((((((....)))))))))..........................((((((((((......)))...((.((((((......)))))).)))))))))..... (-19.94 = -20.54 + 0.60)

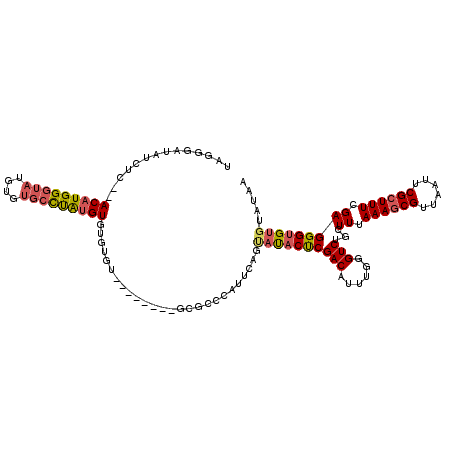
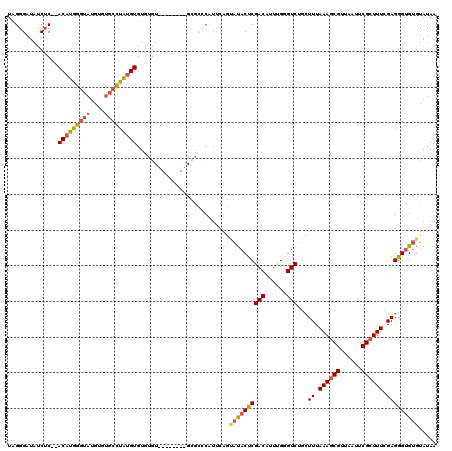
| Location | 6,624,921 – 6,625,033 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.05 |
| Mean single sequence MFE | -26.65 |
| Consensus MFE | -17.84 |
| Energy contribution | -20.44 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6624921 112 - 23771897 AAUAAUCAAUUCGGUAGUCACUCAAUUAUUGAGCCCUUCAUUAUACACACCCUCGAAAGCGAAUUAACGCUUUAAAGCAGACCCAAAUGUCGAGUAUACUGAAUGGGCGC--------AC ..........((((((((......))))))))((((((((.(((((.....((..((((((......))))))..))..(((......)))..))))).)))).))))..--------.. ( -30.20) >DroSec_CAF1 1957 112 - 1 AAUAAUCAAUUCGGUAGUCACUCAAUUAUUGAGCCCUUCAUUAUACACACCCUCGAAAGCGAAUUAACGCUUUAAAGCAGACCCAAAUGUCGAGUAUACUGAAUGGGCGC--------AC ..........((((((((......))))))))((((((((.(((((.....((..((((((......))))))..))..(((......)))..))))).)))).))))..--------.. ( -30.20) >DroSim_CAF1 10098 112 - 1 AAUAAUCAAUUCGGUAGUCACUCAAUUAUUGAGCCCUUCAUUAUACACACCCUCGAAAGCGAAUUAACGCUUUAAAGCAGACCCAAAUGUCGAGUAUACUGAAUGGGCGC--------AC ..........((((((((......))))))))((((((((.(((((.....((..((((((......))))))..))..(((......)))..))))).)))).))))..--------.. ( -30.20) >DroYak_CAF1 3992 97 - 1 AAUAAUCAAUUCGGUAGUCACUCAAUUAUUGAGCCCUUCAUUAUACACACCCUCGAAACCGAAUUAACGCUUUAAAGCAGACCCAAAUGUCGAGUA-----------------------C .......((((((((.....((((.....))))...(((...............)))))))))))...(((....))).(((......))).....-----------------------. ( -14.16) >DroAna_CAF1 2196 117 - 1 AAUAAUCAAUUCGGUAUUCACUCAAUUAUUGAGCCCUUCAUUAUACACACCCUCAAAAGCGAAUUAACGCUUUAAA-CUGACCCA-AUGUCGGGAGUG-UGGAUGCCCACCACAUCCUAU ............(((((((.((((.....))))...........((((.(((...((((((......))))))...-..(((...-..)))))).)))-))))))))............. ( -28.50) >consensus AAUAAUCAAUUCGGUAGUCACUCAAUUAUUGAGCCCUUCAUUAUACACACCCUCGAAAGCGAAUUAACGCUUUAAAGCAGACCCAAAUGUCGAGUAUACUGAAUGGGCGC________AC ..........((((((((......))))))))((((((((.(((((.........((((((......))))))......(((......)))..))))).)))).))))............ (-17.84 = -20.44 + 2.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:30 2006