
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,604,084 – 6,604,233 |
| Length | 149 |
| Max. P | 0.925736 |

| Location | 6,604,084 – 6,604,180 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -16.83 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6604084 96 + 23771897 CAUUAUUAUUAUUUUGCCUUUUU----GGGCCAUUUUUUAAUAUUUGCCUCGG-CGCCGAAAAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGA ...(((((..(..(.((((....----)))).)..)..)))))..((((.(((-((((........))).(((...)))))))..))))............ ( -22.90) >DroVir_CAF1 10536 83 + 1 UAU--------------UUUUUU----UGGACGUUUUUUAAUAUUUGCGCCGUUUGCCAAAAAGUAGGCCGACAUUGUUGCCGCUGGCAUACGGCGCAGGA ...--------------......----................((((((((((.(((((....((.(((.(((...))))))))))))).)))))))))). ( -28.40) >DroPse_CAF1 6476 96 + 1 CAUUAUUAUUAUUUUCCCUUUUU----GGGCCAUUUUUUAAUAUUUGCCACGG-CGCCGAAAAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGA ...(((((.......(((.....----)))........)))))..((((((((-((((........))).(((...))))))).)))))............ ( -22.96) >DroMoj_CAF1 5387 85 + 1 UCU------------CUUUUUUU----CGGGUGUUUUUUAAUAUUUGCGCCGUUUGCCGAAAAGUAGGCCGACAUUGUUGCCGCUGGCAUACGGCGCAGGA .((------------(.......----.)))............((((((((((.((((....(((.(((.(((...))))))))))))).)))))))))). ( -28.50) >DroAna_CAF1 5292 99 + 1 CAUUAUUAUUAUUUUCCUUCUUUUUCGGGGCCAUUUUUUAAUAUUUGCCUCGA-CGCCGA-AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGA ...............(((.((((((((((((...............)))))))-....))-))).)))((..((.((.((((...))))..)).))..)). ( -22.26) >DroPer_CAF1 5703 96 + 1 CAUUAUUAUUAUUUUCCCUUUUU----GGGCCAUUUUUUAAUAUUUGCCACGG-CGCCGAAAAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGA ...(((((.......(((.....----)))........)))))..((((((((-((((........))).(((...))))))).)))))............ ( -22.96) >consensus CAUUAUUAUUAUUUUCCCUUUUU____GGGCCAUUUUUUAAUAUUUGCCACGG_CGCCGAAAAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGA ...........................................(((((.((((..(((....(((.(((.(((...))))))))))))..)))).))))). (-16.83 = -16.42 + -0.42)



| Location | 6,604,084 – 6,604,180 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -23.19 |
| Consensus MFE | -14.82 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6604084 96 - 23771897 UCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUUUUCGGCG-CCGAGGCAAAUAUUAAAAAAUGGCCC----AAAAAGGCAAAAUAAUAAUAAUG ............((((...(....)....(((((((.......)).)-))))))))..(((((.......(((.----.....))).......)))))... ( -21.04) >DroVir_CAF1 10536 83 - 1 UCCUGCGCCGUAUGCCAGCGGCAACAAUGUCGGCCUACUUUUUGGCAAACGGCGCAAAUAUUAAAAAACGUCCA----AAAAAA--------------AUA ...((((((((.(((((((((((....))))).........)))))).))))))))..................----......--------------... ( -27.30) >DroPse_CAF1 6476 96 - 1 UCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUUUUCGGCG-CCGUGGCAAAUAUUAAAAAAUGGCCC----AAAAAGGGAAAAUAAUAAUAAUG ............(((((.((((..(......)(((........))))-))))))))..(((((........(((----.....))).......)))))... ( -23.16) >DroMoj_CAF1 5387 85 - 1 UCCUGCGCCGUAUGCCAGCGGCAACAAUGUCGGCCUACUUUUCGGCAAACGGCGCAAAUAUUAAAAAACACCCG----AAAAAAAG------------AGA ...((((((((.((((..(((((....)))))(....).....)))).))))))))..................----........------------... ( -25.50) >DroAna_CAF1 5292 99 - 1 UCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU-UCGGCG-UCGAGGCAAAUAUUAAAAAAUGGCCCCGAAAAAGAAGGAAAAUAAUAAUAAUG (((((((.....)))...(((((....)))))(((.....-..))).-(((.(((...............))).)))......)))).............. ( -18.96) >DroPer_CAF1 5703 96 - 1 UCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUUUUCGGCG-CCGUGGCAAAUAUUAAAAAAUGGCCC----AAAAAGGGAAAAUAAUAAUAAUG ............(((((.((((..(......)(((........))))-))))))))..(((((........(((----.....))).......)))))... ( -23.16) >consensus UCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUUUUCGGCG_CCGAGGCAAAUAUUAAAAAAUGGCCC____AAAAAAGGAAAAUAAUAAUAAUG ...(((.(((........(((((....)))))(((........)))...))).)))............................................. (-14.82 = -14.93 + 0.11)
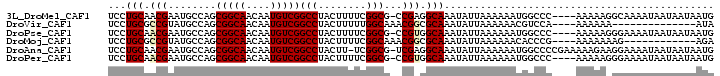


| Location | 6,604,140 – 6,604,233 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6604140 93 + 23771897 AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGAAAAGGAAAUGCCGCAGCAGCACUUGCCUUGCUCUGCAGAGUAGAGCAGGCCCU------------ .....(((((.((.((((((.((.(((((((.((.......)).).)))))))).))))))..)).).((((((((...))))))))))))..------------ ( -38.20) >DroPse_CAF1 6532 91 + 1 AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGAAAAGGAAAUGCCGCAGUAGCAGCAG----------UAGC----ACGAGUGGCAGGAGUGUGGCAG ......(((.(((((.(((((((((((((((.((.......)).).))))))((....)).((..----------..))----...)))))))).)))))))).. ( -35.40) >DroEre_CAF1 3805 93 + 1 AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGAAAAGGAAAUGCCGCAGCAGCACUUGCCUCGCUCUGCAGCGUAGAGAAGGCCCU------------ .....(((((.((.((((((.((.(((((((.((.......)).).)))))))).))))))..)).)...((((((...))))))..))))..------------ ( -33.50) >DroWil_CAF1 23461 71 + 1 AAGUAGGCCGACAUUGUUCCCUUUGGCAUUUGUUGCAGGAAAAGGAAAUACAGCAACAGCAGCAG----------CAGC----UG-------------------- ......(((((...........)))))..(((((((.(............).)))))))((((..----------..))----))-------------------- ( -14.50) >DroAna_CAF1 5351 79 + 1 AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGAAAAGGAAAUGCCGCAGCAGCAGAAG----------CAGA----AGCAGUAGCA------------ ......((..((.(((((((.((.(((((((.((.......)).).)))))))).)))))))..(----------(...----.)).)).)).------------ ( -23.70) >DroPer_CAF1 5759 91 + 1 AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGAAAAGGAAAUGCCGCAGUAGCAGCAG----------UAGC----ACGAGUGGCAAGAGUGUGGCAG ......(((.(((((.(((((((((((((((.((.......)).).))))))((....)).((..----------..))----...)))))))).)))))))).. ( -35.70) >consensus AAGUAGGCCGACAUUGUUGCCGCUGGCAUUCGUUGCAGGAAAAGGAAAUGCCGCAGCAGCAGCAG__________CAGC____AGCAGUCGCA____________ ..............((((((.((.(((((((.((.......)).).)))))))).))))))............................................ (-14.59 = -14.98 + 0.39)


| Location | 6,604,140 – 6,604,233 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -10.58 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6604140 93 - 23771897 ------------AGGGCCUGCUCUACUCUGCAGAGCAAGGCAAGUGCUGCUGCGGCAUUUCCUUUUCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU ------------.(((((((((((.......)))))).((((..((.(((((((((((((..............))))))).)))))).)).))))))))).... ( -38.64) >DroPse_CAF1 6532 91 - 1 CUGCCACACUCCUGCCACUCGU----GCUA----------CUGCUGCUACUGCGGCAUUUCCUUUUCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU ..(((......(((.((.((((----((..----------.((((((....))))))............)).)))).)).))).((((....)))))))...... ( -24.16) >DroEre_CAF1 3805 93 - 1 ------------AGGGCCUUCUCUACGCUGCAGAGCGAGGCAAGUGCUGCUGCGGCAUUUCCUUUUCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU ------------.(((((.......((((....)))).((((..((.(((((((((((((..............))))))).)))))).)).))))))))).... ( -36.34) >DroWil_CAF1 23461 71 - 1 --------------------CA----GCUG----------CUGCUGCUGUUGCUGUAUUUCCUUUUCCUGCAACAAAUGCCAAAGGGAACAAUGUCGGCCUACUU --------------------((----((..----------..))))..((.(((((((((((((((...(((.....))).)))))))..)))).))))..)).. ( -17.50) >DroAna_CAF1 5351 79 - 1 ------------UGCUACUGCU----UCUG----------CUUCUGCUGCUGCGGCAUUUCCUUUUCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU ------------.(((((.((.----...)----------)...((.(((((((((((((..............))))))).)))))).))..)).)))...... ( -21.04) >DroPer_CAF1 5759 91 - 1 CUGCCACACUCUUGCCACUCGU----GCUA----------CUGCUGCUACUGCGGCAUUUCCUUUUCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU ..((((((.(.(((((....((----((..----------.....)).)).(((((((((..............))))))).))))))).).))).)))...... ( -24.34) >consensus ____________UGCCACUCCU____GCUG__________CUGCUGCUGCUGCGGCAUUUCCUUUUCCUGCAACGAAUGCCAGCGGCAACAAUGUCGGCCUACUU .............................................(((...(((((((((..............))))))).))((((....)))))))...... (-10.58 = -10.97 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:14 2006