
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,600,026 – 6,600,139 |
| Length | 113 |
| Max. P | 0.999859 |

| Location | 6,600,026 – 6,600,139 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -33.96 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6600026 113 + 23771897 GUGCUACCUG--UUCAGGUACCAUGCAAAAGCGCAGCACAAAAUUGCUGCAUCAACACUUCGUUUGCGCUUUUUGUUGUUGCCUGUCGGGGCGGCUUUUCUCGCUACUUGCUAGU--- ..(((.((((--..((((((.((.((((((((((((((......))))))...(((.....)))...)).)))))))).)))))).))))..)))....((.((.....)).)).--- ( -36.40) >DroSec_CAF1 62980 113 + 1 GUGCUACCUG--UUCAGCUGCCAUGCAAAAGCGCAGCACAAAAUUGCUGCAUCAACACUUCGUUUGCGCUUUUUGUUGUUGCCUGUCGGGGCGGCUUUUCUCGCUACUUGCUAGU--- ..((((....--...(((((((.......((.(((((((((((..((.(((..(((.....)))))))).))))).)))))))).....)))))))......((.....))))))--- ( -32.00) >DroSim_CAF1 63165 113 + 1 GUGCUACCUG--UUCAGGUGCCAUGCAAAAGCGCAGCACAAAAUUGCUGCAUCAACACUUCGUUUGCGCUUUUUGUUGUUGCCUGUCGGGGCGGCUUUUCUCGCUACUUGCUAGU--- ..(((.((((--..(((((..((.((((((((((((((......))))))...(((.....)))...)).))))))))..))))).))))..)))....((.((.....)).)).--- ( -36.10) >DroEre_CAF1 63238 113 + 1 GUGCUACCUG--UUCAGGUGGCAUGCAAAAGCGCAGCACAAAAUUGCUGCAUCAACACUUCGUUUGCGCUUUUUGUUGUUGCCUGUCGGGGCGGCUUUUCUCGCUGCUUGCUAGU--- ..((..((((--..((((..(((.((((((((((((((......))))))...(((.....)))...)).)))))))))..)))).))))(((((.......)))))..))....--- ( -43.60) >DroYak_CAF1 64876 118 + 1 GUGCUACCUGUGUUCAGGUGCCAUGCAAAAGCGCAGCACAAAAUUGCUGCAUCAACACUUCGUUUGCGCUUUUUGUUGUUGCCUGUCGGGGCGGCUUUCCUCGCUACUUGCAGGUGUG (((.((((((....)))))).)))((((((((((((((......))))))...(((.....)))...)).))))))....((((((.(((((((......))))).)).))))))... ( -39.40) >consensus GUGCUACCUG__UUCAGGUGCCAUGCAAAAGCGCAGCACAAAAUUGCUGCAUCAACACUUCGUUUGCGCUUUUUGUUGUUGCCUGUCGGGGCGGCUUUUCUCGCUACUUGCUAGU___ ..(((.((((....(((((..((.((((((((((((((......))))))...(((.....)))...)).))))))))..))))).))))..)))....((.((.....)).)).... (-33.96 = -34.00 + 0.04)



| Location | 6,600,026 – 6,600,139 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -36.86 |
| Consensus MFE | -34.92 |
| Energy contribution | -35.16 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.28 |
| SVM RNA-class probability | 0.999859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6600026 113 - 23771897 ---ACUAGCAAGUAGCGAGAAAAGCCGCCCCGACAGGCAACAACAAAAAGCGCAAACGAAGUGUUGAUGCAGCAAUUUUGUGCUGCGCUUUUGCAUGGUACCUGAA--CAGGUAGCAC ---.((.((.....)).))....((..((....((((...((...(((((((((.(((((((((((...)))).)))))))..)))))))))...))...))))..--..))..)).. ( -36.10) >DroSec_CAF1 62980 113 - 1 ---ACUAGCAAGUAGCGAGAAAAGCCGCCCCGACAGGCAACAACAAAAAGCGCAAACGAAGUGUUGAUGCAGCAAUUUUGUGCUGCGCUUUUGCAUGGCAGCUGAA--CAGGUAGCAC ---.((.((.....)).))....((((((......))).......(((((((((.(((((((((((...)))).)))))))..)))))))))....))).((((..--....)))).. ( -35.30) >DroSim_CAF1 63165 113 - 1 ---ACUAGCAAGUAGCGAGAAAAGCCGCCCCGACAGGCAACAACAAAAAGCGCAAACGAAGUGUUGAUGCAGCAAUUUUGUGCUGCGCUUUUGCAUGGCACCUGAA--CAGGUAGCAC ---...........((.......((((((......))).......(((((((((.(((((((((((...)))).)))))))..)))))))))....)))((((...--.)))).)).. ( -36.40) >DroEre_CAF1 63238 113 - 1 ---ACUAGCAAGCAGCGAGAAAAGCCGCCCCGACAGGCAACAACAAAAAGCGCAAACGAAGUGUUGAUGCAGCAAUUUUGUGCUGCGCUUUUGCAUGCCACCUGAA--CAGGUAGCAC ---....((..(((((.......(((.........))).......(((((((((.(((((((((((...)))).)))))))..))))))))))).))).((((...--.)))).)).. ( -36.10) >DroYak_CAF1 64876 118 - 1 CACACCUGCAAGUAGCGAGGAAAGCCGCCCCGACAGGCAACAACAAAAAGCGCAAACGAAGUGUUGAUGCAGCAAUUUUGUGCUGCGCUUUUGCAUGGCACCUGAACACAGGUAGCAC ....(((((.....)).)))...((((((......))).......(((((((((.(((((((((((...)))).)))))))..)))))))))....)))(((((....)))))..... ( -40.40) >consensus ___ACUAGCAAGUAGCGAGAAAAGCCGCCCCGACAGGCAACAACAAAAAGCGCAAACGAAGUGUUGAUGCAGCAAUUUUGUGCUGCGCUUUUGCAUGGCACCUGAA__CAGGUAGCAC ..............((.......((((((......))).......(((((((((.(((((((((((...)))).)))))))..)))))))))....)))(((((....))))).)).. (-34.92 = -35.16 + 0.24)


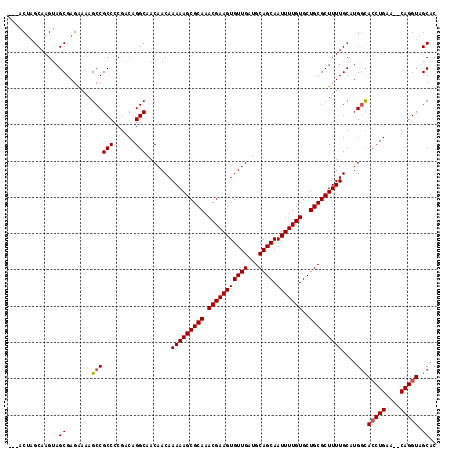
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:10 2006