| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 640,108 – 640,228 |
| Length | 120 |
| Max. P | 0.564182 |

| Location | 640,108 – 640,228 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -27.17 |
| Energy contribution | -26.65 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
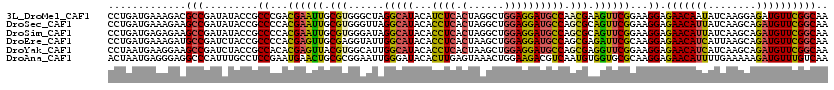
>3L_DroMel_CAF1 640108 120 + 23771897 CCUGAUGAAAGACGCCGAUAUACCGCCCGACGAAUUGCGUGGGCUAGGCAUACAUCUCACUAGGCUGGAGGAUGCCAACGAAGUUCGGAAGGAGAACAAUAUCAAGGAGAUGUUCGGCAA .............(((((......((((.(((.....))))))).......(((((((.((.(((........)))......((((.......)))).......)))))))))))))).. ( -37.60) >DroSec_CAF1 7395 120 + 1 CCUGAUGAAAGAAGCCGAUAUACCGCCCCACGAAUUGCGUGGGUUAGGCAUACACCUCACUAGGCUGGAGGAUGCCAGCGCAGUUCGGAAGGAGAACAUUAUCAAGCAGAUGUUCGGCAA .............(((.........((...((((((((((......(((((...((((.(......)))))))))).))))))))))...)).(((((((........)))))))))).. ( -40.40) >DroSim_CAF1 9890 120 + 1 CCUGAUGAGAGAAGCCGAUAUACCGCCCCACGAAUUGCGUGGGAUAGGCAUACACCUCACUAGGCUGGAGGAUGCCAGCGCAGUUCGGAAGGAGAACAUUAUCAAGCAGAUGUUCGGCAA .............(((.......(((((((((.....))))))...(((((...((((.(......)))))))))).)))...(((....)))(((((((........)))))))))).. ( -40.60) >DroEre_CAF1 10262 120 + 1 CCUGAUGAAAGAUGCCGAUCUACCGCCCCACGAGUUGCGAGGUAUUGGCAUACACCUCACUAAGCUGGAGGAUGCCAGCGAGAUUCGCAAGGAGAACAUCAUUAAGCAGAUGUUCGGCAA ...........((((((((((..(((..........))).)).))))))))...((((.(......))))).((((.(((.....))).....(((((((........))))))))))). ( -37.50) >DroYak_CAF1 7557 120 + 1 CCUAAUGAAGGAAGCCGAUCUACCGCCACACGAGUUACGUGGCAUUGGCAUACACCUCACUAAGCUGGAGGAUGCCAGCGAGGUUCGGAAGGAGAACAUCAUCAAGCAGAUGUUCGGCAA (((.....)))..(((......(((((((.........))))).(((((((...((((.(......))))))))))))...))(((....)))(((((((........)))))))))).. ( -41.70) >DroAna_CAF1 7100 120 + 1 ACUAAUGAGGGAGGCCCAUUUGCCUCCGAAUGAACUGCGCGGAAUUGGGAUACACUUGAGUAAACUGGAAGACGUCAAUGUGGUGCGCAAGGAGAACAUUUUGAAAAAGAUGUUUGUCAA .........((((((......))))))........((((((..((...(((....((.((....)).))....)))...))..)))))).(..(((((((((....)))))))))..).. ( -33.90) >consensus CCUGAUGAAAGAAGCCGAUAUACCGCCCCACGAAUUGCGUGGGAUAGGCAUACACCUCACUAAGCUGGAGGAUGCCAGCGAAGUUCGGAAGGAGAACAUUAUCAAGCAGAUGUUCGGCAA .............(((.........((...((((((.(((......(((((...((((.(......)))))))))).))).))))))...)).(((((((........)))))))))).. (-27.17 = -26.65 + -0.52)

| Location | 640,108 – 640,228 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -24.24 |
| Energy contribution | -25.60 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
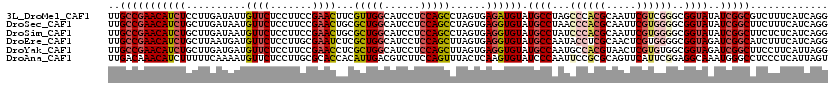
>3L_DroMel_CAF1 640108 120 - 23771897 UUGCCGAACAUCUCCUUGAUAUUGUUCUCCUUCCGAACUUCGUUGGCAUCCUCCAGCCUAGUGAGAUGUAUGCCUAGCCCACGCAAUUCGUCGGGCGGUAUAUCGGCGUCUUUCAUCAGG ..((((((((((((((.(.....((((.......))))...(((((......)))))).)).)))))))(((((..(((((((.....))).))))))))).)))))............. ( -39.50) >DroSec_CAF1 7395 120 - 1 UUGCCGAACAUCUGCUUGAUAAUGUUCUCCUUCCGAACUGCGCUGGCAUCCUCCAGCCUAGUGAGGUGUAUGCCUAACCCACGCAAUUCGUGGGGCGGUAUAUCGGCUUCUUUCAUCAGG ..((((((((((((((.(.....((((.......))))...(((((......)))))).))).))))))(((((...((((((.....))))))..))))).)))))............. ( -40.60) >DroSim_CAF1 9890 120 - 1 UUGCCGAACAUCUGCUUGAUAAUGUUCUCCUUCCGAACUGCGCUGGCAUCCUCCAGCCUAGUGAGGUGUAUGCCUAUCCCACGCAAUUCGUGGGGCGGUAUAUCGGCUUCUCUCAUCAGG ..((((((((((((((.(.....((((.......))))...(((((......)))))).))).))))))(((((..(((((((.....))))))).))))).)))))............. ( -40.00) >DroEre_CAF1 10262 120 - 1 UUGCCGAACAUCUGCUUAAUGAUGUUCUCCUUGCGAAUCUCGCUGGCAUCCUCCAGCUUAGUGAGGUGUAUGCCAAUACCUCGCAACUCGUGGGGCGGUAGAUCGGCAUCUUUCAUCAGG .(((((((((((........))))))).(((..(((.....(((((......)))))...(((((((((......)))))))))...)))..))).........))))............ ( -46.70) >DroYak_CAF1 7557 120 - 1 UUGCCGAACAUCUGCUUGAUGAUGUUCUCCUUCCGAACCUCGCUGGCAUCCUCCAGCUUAGUGAGGUGUAUGCCAAUGCCACGUAACUCGUGUGGCGGUAGAUCGGCUUCCUUCAUUAGG ..((((((((((........)))))))..((.((..(((((((((((........)).)))))))))....((((....((((.....)))))))))).))...)))..(((.....))) ( -42.20) >DroAna_CAF1 7100 120 - 1 UUGACAAACAUCUUUUUCAAAAUGUUCUCCUUGCGCACCACAUUGACGUCUUCCAGUUUACUCAAGUGUAUCCCAAUUCCGCGCAGUUCAUUCGGAGGCAAAUGGGCCUCCCUCAUUAGU ((((.(((....))).))))((((......((((((.....((((........)))).(((......)))..........)))))).......((((((......))))))..))))... ( -22.50) >consensus UUGCCGAACAUCUGCUUGAUAAUGUUCUCCUUCCGAACCUCGCUGGCAUCCUCCAGCCUAGUGAGGUGUAUGCCAAUCCCACGCAAUUCGUGGGGCGGUAUAUCGGCUUCUUUCAUCAGG ..(((((((((((..........((((.......))))...(((((......)))))......)))))).((((...((((((.....))))))..))))..)))))............. (-24.24 = -25.60 + 1.36)
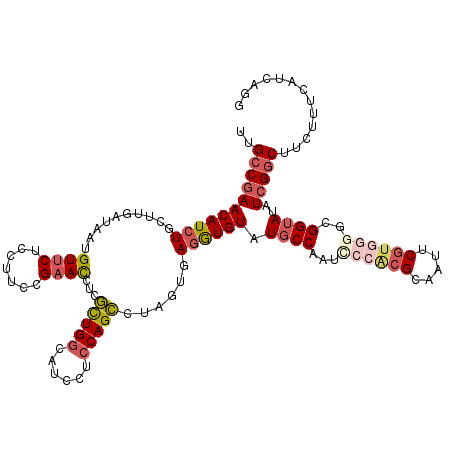
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:01 2006