
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,555,648 – 6,555,768 |
| Length | 120 |
| Max. P | 0.999737 |

| Location | 6,555,648 – 6,555,768 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.49 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -10.15 |
| Energy contribution | -15.65 |
| Covariance contribution | 5.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
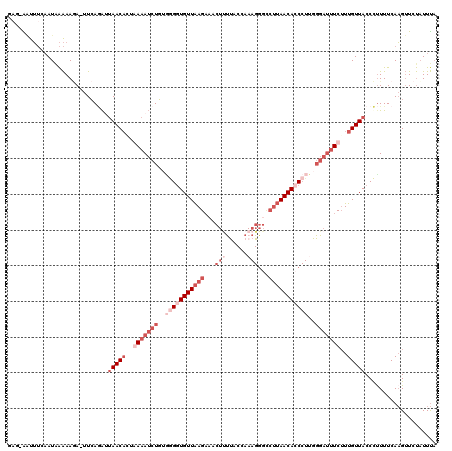
>3L_DroMel_CAF1 6555648 120 + 23771897 GAGAGAUUUCAAUAAAAAAAGUUGAAACUAACAGUGAAAUCUCUGAGGUGUUAAGCAACUUUUACGAAAGGGCCUUAACACCCUUGGGAUUUCUUUGUUACCUCUUUCAAGUUCUAUUUA (((((((((((((.......))))))).((((((.((((((((...(((((((((...((((....))))...)))))))))...)))))))).))))))..))))))............ ( -38.60) >DroSec_CAF1 18365 118 + 1 GAG-AAUUUCAAUGAAAAUA-UUCAUAUGAACUCUAAAAUCUGUGGGGUGUUAAGAUACUUCCACCAAAGGGUCUUAACACCCUUGGGAUUUCUUUGUUACCCUUUUCAAGUUCUAUUUA .((-(((((..(((((....-)))))...(((....((((((..((((((((((((..(((......)))..))))))))))))..))))))....))).........)))))))..... ( -33.50) >DroSim_CAF1 18434 115 + 1 ----AGUUUCAAUAAAAAGA-UUCAGAUUAACACUAAAAUCUGGUGGGUGUUAAGAUACUCUUACCAAAGGGCCUUAACAACCUAGGGAUUUCUUUGUUACCCUUUUCAAGUUCUUUUUA ----........((((((((-.(((((((........))))))).(((((.((((.....)))).((((((.(((((......)))))...)))))).))))).........)))))))) ( -26.40) >DroEre_CAF1 18561 94 + 1 AAG-G-UAAACAUAGGAAGA-UUCAGGAUAACACUGAAAACUGUAGGAUGUUGAGAAACUA------A--------AACACC---------UCGUUGUUACCCUUUUCAUGAUCCAUUCU ..(-(-....(((..((((.-(((((.......)))))(((...(((.((((.........------.--------))))))---------)....)))...))))..)))..))..... ( -16.60) >consensus GAG_AAUUUCAAUAAAAAGA_UUCAGAUUAACACUAAAAUCUGUGGGGUGUUAAGAAACUUUUACCAAAGGGCCUUAACACCCUUGGGAUUUCUUUGUUACCCUUUUCAAGUUCUAUUUA ............................(((((..(((((((..(((((((((((...(((......)))...)))))))))))..)))))))..))))).................... (-10.15 = -15.65 + 5.50)


| Location | 6,555,648 – 6,555,768 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.49 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -16.75 |
| Energy contribution | -20.56 |
| Covariance contribution | 3.81 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
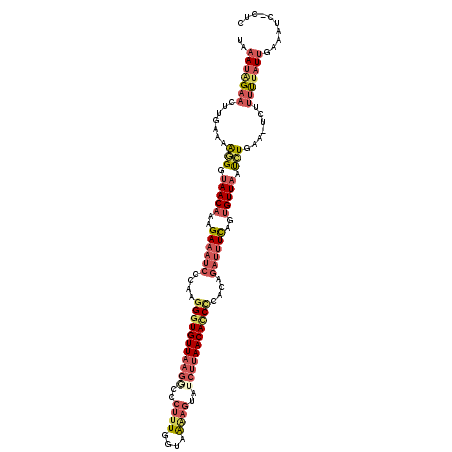
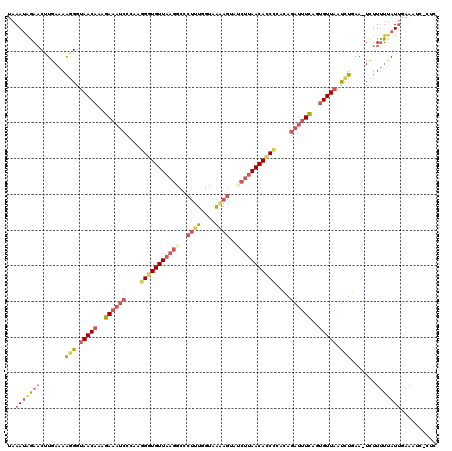
>3L_DroMel_CAF1 6555648 120 - 23771897 UAAAUAGAACUUGAAAGAGGUAACAAAGAAAUCCCAAGGGUGUUAAGGCCCUUUCGUAAAAGUUGCUUAACACCUCAGAGAUUUCACUGUUAGUUUCAACUUUUUUUAUUGAAAUCUCUC ...............((((.(((((..((((((.(..((((((((((...((((....))))...))))))))))..).))))))..)))))(((((((.........))))))))))). ( -30.00) >DroSec_CAF1 18365 118 - 1 UAAAUAGAACUUGAAAAGGGUAACAAAGAAAUCCCAAGGGUGUUAAGACCCUUUGGUGGAAGUAUCUUAACACCCCACAGAUUUUAGAGUUCAUAUGAA-UAUUUUCAUUGAAAUU-CUC ......((((((.(((((((............)))..(((((((((((..((((....))))..)))))))))))......)))).))))))..(((((-....))))).......-... ( -33.00) >DroSim_CAF1 18434 115 - 1 UAAAAAGAACUUGAAAAGGGUAACAAAGAAAUCCCUAGGUUGUUAAGGCCCUUUGGUAAGAGUAUCUUAACACCCACCAGAUUUUAGUGUUAAUCUGAA-UCUUUUUAUUGAAACU---- ((((((((.(((....))).(((((..((((((....((.((((((((..((((....))))..)))))))).))....))))))..))))).......-))))))))........---- ( -29.50) >DroEre_CAF1 18561 94 - 1 AGAAUGGAUCAUGAAAAGGGUAACAACGA---------GGUGUU--------U------UAGUUUCUCAACAUCCUACAGUUUUCAGUGUUAUCCUGAA-UCUUCCUAUGUUUA-C-CUU .....(((..((....(((((((((((.(---------((((((--------.------.........)))).)))...))......)))))))))..)-)..)))........-.-... ( -17.10) >consensus UAAAUAGAACUUGAAAAGGGUAACAAAGAAAUCCCAAGGGUGUUAAGGCCCUUUGGUAAAAGUAUCUUAACACCCCACAGAUUUCAGUGUUAAUCUGAA_UCUUUUUAUUGAAAUC_CUC ..(((((((.......(((.(((((..((((((....(((((((((((..((((....))))..)))))))))))....))))))..))))).))).......))))))).......... (-16.75 = -20.56 + 3.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:51 2006