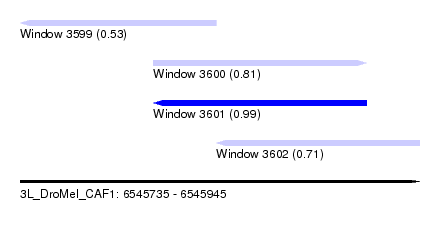
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,545,735 – 6,545,945 |
| Length | 210 |
| Max. P | 0.994072 |

| Location | 6,545,735 – 6,545,838 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -20.25 |
| Consensus MFE | -13.52 |
| Energy contribution | -14.02 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6545735 103 - 23771897 GCUGCACCACCACCACAGCAAUCAGGUGAAUCAGCACCACCACCAGCAGCAGCAGAGUCACCACCAUUUGCAGCACGC------CAACUUGCACUCCCACCAUCCACAC (((((...........(....)..((((......)))).......))))).(((((..........))))).(((...------.....)))................. ( -18.20) >DroSec_CAF1 8288 100 - 1 GCUACACCACCACCACAGCAGCCAGGUGAAUCAGCACC---ACCAGCAGCAGCAGAGUCACCACCAUUUGCAGCACGC------CAGCUCGCACUCCCACCAUCCACAC .................(((((..((((.....((...---....)).((.(((((..........))))).)).)))------).))).))................. ( -18.30) >DroSim_CAF1 8376 100 - 1 GCUGCACCACCACCACAGCAGCCAGGUGAAUCAGCACC---ACCAGCAGCAGCAGAGUCACCACCAUUUGCAGCACGC------CAGCUCGCACUCCCACCAUCCACAC (((((............)))))..((((.....((...---....)).((.(((((..........)))))(((....------..))).)).....))))........ ( -21.90) >DroEre_CAF1 8324 103 - 1 GCUGCACCACCACCACGCCAACCAGGUGAAUCCGCACC---AC---CAGCAGCAGAGCCACCACCAUUUGCAGCACUCGAGCUCGAACUCGAACUCCCACCAUCAACAC ((((((..........(.....).((((..((.((...---..---.....)).))..))))......))))))....(((.(((....))).)))............. ( -22.60) >consensus GCUGCACCACCACCACAGCAACCAGGUGAAUCAGCACC___ACCAGCAGCAGCAGAGUCACCACCAUUUGCAGCACGC______CAACUCGCACUCCCACCAUCCACAC ((((...((((.............))))...)))).............((.(((((..........))))).))................................... (-13.52 = -14.02 + 0.50)

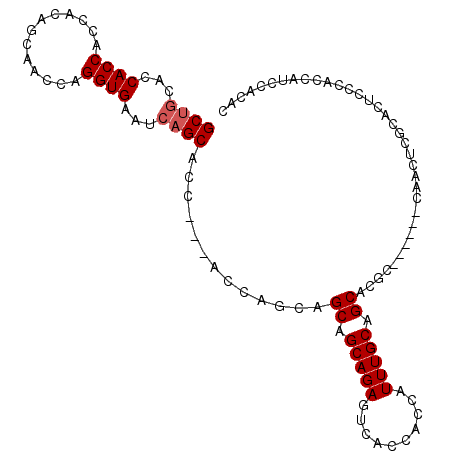
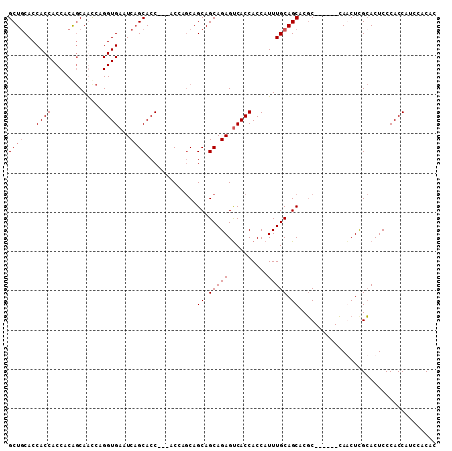
| Location | 6,545,805 – 6,545,917 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 72.38 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -23.43 |
| Energy contribution | -23.35 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6545805 112 + 23771897 UGAUUCACCUGAUUGCUGU-GGUGGUGGUGCAGC---UGC-UGCGAGUGGCGACCUAAGCCAGUUCCAGCUGCAUCAUGACAGCUGGUGGCCAAGUCGAGAUGGGAAACGGAUUUGU ....(((((.....(((((-....((((((((((---((.-.((...((((.......))))))..)))))))))))).))))).))))).((((((......(....).)))))). ( -41.80) >DroVir_CAF1 20743 103 + 1 G----------GGUGUUGCGUCUGAUGGUGCAGU---UGCCUGUG-CCUGUGAUCUAUGCCAGCGCCAGCUGCGCCAUGGCAGCUGGUGGCCAAAUCGAGAUGCGAAACGGAUUUGU .----------.((.(((((((((((((..(((.---...)))..-))..........(((...(((((((((......))))))))))))...))).)))))))).))........ ( -43.30) >DroGri_CAF1 20196 97 + 1 U----------GCA------GUUGCACUUGCAGU---UGCCUGCG-UUGCCGAUCUAUGCCAGCGCCAGCUGCACCAUGGCAGCUGGUGGCCAAAUCGAGAUGCGAUACGGAUUUGU .----------(((------..(((....)))..---))).((((-((..((((....(((...(((((((((......))))))))))))...)))).))))))............ ( -38.50) >DroEre_CAF1 8394 112 + 1 GGAUUCACCUGGUUGGCGU-GGUGGUGGUGCAGC---UGC-UGCGAGUGGCGACCUAAGCCAGUUCCAGCUGCGUCAUGGCAGCUGGUGGCCAAGUCGAGAUGGGAAACGGAUUUGU ((((((..((.(((((((.-.(..((......))---..)-..)((.((((.......)))).))((((((((......))))))))..)))))..).))...(....))))))).. ( -43.20) >DroAna_CAF1 19923 99 + 1 -----------GCA------GGUGCUGGUGCGACUGGUGG-UGCGGGUGGCGACCUAAGCCAGAACUAGCUGCUUCAUGGCAGCUGGUGGCCAAGUCGAGAUGCGAGACGGAUUUGU -----------(((------(((.((.((.((((((((((-(..((((....))))..)))...((((((((((....)))))))))).))).)))))....)).))....)))))) ( -40.30) >DroPer_CAF1 41339 106 + 1 GGAUGCGAGUGGUGGCCGU-GUGGGUGGU---------GC-UGCGUGUGGCGACCUAUGCCAUUGCCAGCUGCUUCAUGGCAGCUGGUGGCCAAAUCGAGAUGCGACACGGAUUUGU .....(((....(((((((-((((((..(---------((-((....)))))))))))))....((((((((((....)))))))))))))))..)))((((.((...)).)))).. ( -45.10) >consensus G__________GUUG__GU_GGUGGUGGUGCAGC___UGC_UGCG_GUGGCGACCUAAGCCAGUGCCAGCUGCAUCAUGGCAGCUGGUGGCCAAAUCGAGAUGCGAAACGGAUUUGU .........................................((((.....((((....((((...((((((((......))))))))))))...))))...))))............ (-23.43 = -23.35 + -0.08)
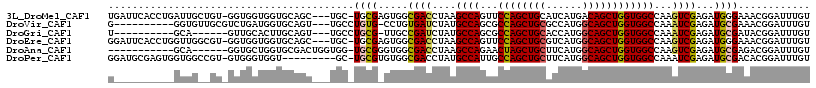

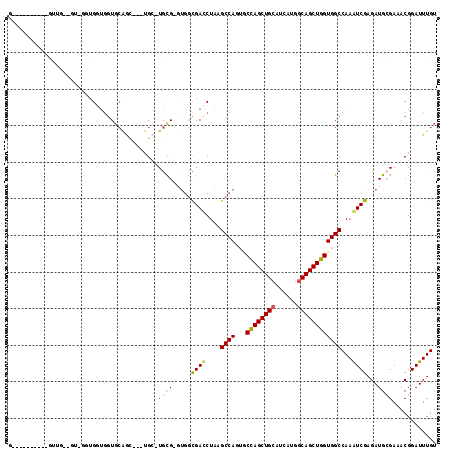
| Location | 6,545,805 – 6,545,917 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.38 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -26.24 |
| Energy contribution | -25.97 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6545805 112 - 23771897 ACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCUGUCAUGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCA---GCUGCACCACCACC-ACAGCAAUCAGGUGAAUCA ........(.(((((....(((((((((((((((((((......))))))))...)))).))))))).........-...---((((..........-.)))).....)).))).). ( -35.20) >DroVir_CAF1 20743 103 - 1 ACAAAUCCGUUUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGGCGCAGCUGGCGCUGGCAUAGAUCACAGG-CACAGGCA---ACUGCACCAUCAGACGCAACACC----------C .............((.(((.((((((((((((((((((......))))))))...)))).))))))...((-..(((...---.)))..))...))).))......----------. ( -37.60) >DroGri_CAF1 20196 97 - 1 ACAAAUCCGUAUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGGUGCAGCUGGCGCUGGCAUAGAUCGGCAA-CGCAGGCA---ACUGCAAGUGCAAC------UGC----------A .............(((...(((((((((((((((((((......))))))))...)))).)))))))(((.-.((((...---.))))...)))...------)))----------. ( -42.80) >DroEre_CAF1 8394 112 - 1 ACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCUGCCAUGACGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCA---GCUGCACCACCACC-ACGCCAACCAGGUGAAUCC ...................(((((((((((((((((((......))))))))...)))).)))))))......(((-(..---.)))).........-.((((.....))))..... ( -37.20) >DroAna_CAF1 19923 99 - 1 ACAAAUCCGUCUCGCAUCUCGACUUGGCCACCAGCUGCCAUGAAGCAGCUAGUUCUGGCUUAGGUCGCCACCCGCA-CCACCAGUCGCACCAGCACC------UGC----------- .............((....((((((((((((.((((((......)))))).))...))).)))))))......)).-....(((..((....))..)------)).----------- ( -27.40) >DroPer_CAF1 41339 106 - 1 ACAAAUCCGUGUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGAAGCAGCUGGCAAUGGCAUAGGUCGCCACACGCA-GC---------ACCACCCAC-ACGGCCACCACUCGCAUCC ......((((((........((((((((((((((((((......))))))))...)))).))))))((........-))---------.......))-))))............... ( -34.20) >consensus ACAAAUCCGUUUCGCAUCUCGACUUGGCCACCAGCUGCCAUGAAGCAGCUGGCACUGGCAUAGGUCGCCAC_CGCA_GCA___GCUGCACCACCACC_AC__CAAC__________C ...................(((((((((((((((((((......))))))))...)))).))))))).................................................. (-26.24 = -25.97 + -0.28)

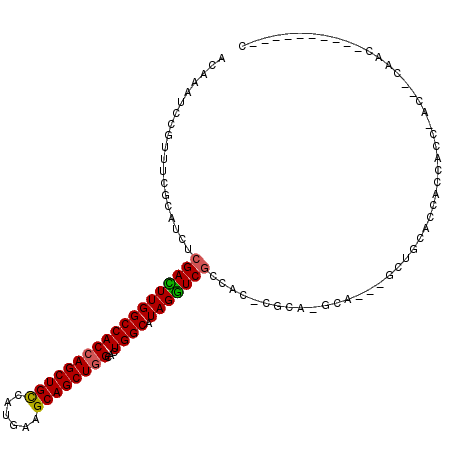
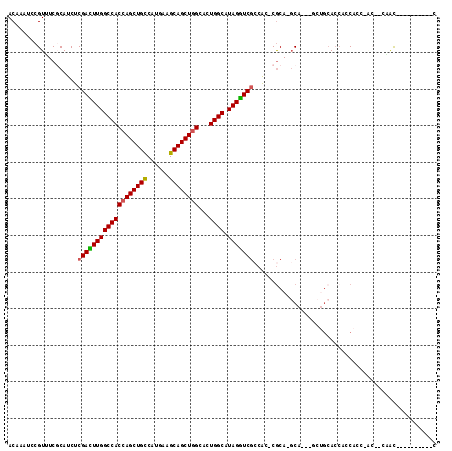
| Location | 6,545,838 – 6,545,945 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -45.34 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.30 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6545838 107 - 23771897 ---UUGAGA------GCCGCUGGCUGGAGACACACGCACAAAUCCGUUUCCCAUCUCGACUUGGCCACCAGCUGUCAUGAUGCAGCUGGAACUGGCUUAGGUCGCCACUCGCA-GCA ---......------((.((((((.((((((..............))))))......((((((((((((((((((......))))))))...)))).))))))))))...)).-)). ( -41.44) >DroVir_CAF1 20767 107 - 1 ---CUGCGC------GCCGCCGGCUGGCGACACACGCACAAAUCCGUUUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGGCGCAGCUGGCGCUGGCAUAGAUCACAGG-CACAGGCA ---......------(((...(.(((((((...(((........))).)))).....((((((((((((((((((......))))))))...)))).)))))).))).-)...))). ( -42.60) >DroGri_CAF1 20214 107 - 1 ---CUGCGA------GCCGCUGGCUGGCGGCACACGCACAAAUCCGUAUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGGUGCAGCUGGCGCUGGCAUAGAUCGGCAA-CGCAGGCA ---(((((.------((((((....))))))....((((......)).........(((((((((((((((((((......))))))))...)))).)))))))))..-)))))... ( -48.70) >DroMoj_CAF1 7493 107 - 1 ---CUGCGC------GCCGCUGGCUGGCGACACACGCACAAAUCCGUCUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGGCGCAGCUGGCGCUGGCAUAGAUCACAGG-CAGAGGCA ---......------(((.(((.(((((((...(((........))).)))).....((((((((((((((((((......))))))))...)))).)))))).))).-))).))). ( -47.80) >DroAna_CAF1 19943 107 - 1 ---UUGAGA------GCCGCCGGCUGGCGACACACCCACAAAUCCGUCUCGCAUCUCGACUUGGCCACCAGCUGCCAUGAAGCAGCUAGUUCUGGCUUAGGUCGCCACCCGCA-CCA ---......------...((.((.(((((((.................(((.....)))((.((((((.((((((......)))))).))...)))).))))))))))).)).-... ( -36.50) >DroPer_CAF1 41367 115 - 1 GGACUGCGGGCUGCUGCCGCUGGCUGGCGGCACACGCACAAAUCCGUGUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGAAGCAGCUGGCAAUGGCAUAGGUCGCCACACGCA-GC- ...((((((((....)))(.((((..(((((((............))))))).....((((((((((((((((((......))))))))...)))).))))))))))).))))-).- ( -55.00) >consensus ___CUGCGA______GCCGCUGGCUGGCGACACACGCACAAAUCCGUCUCGCAUCUCGAUUUGGCCACCAGCUGCCAUGAAGCAGCUGGCGCUGGCAUAGAUCGCCAC_CGCA_GCA ...............(((...)))..((((.((............)).)))).....((((((((((((((((((......))))))))...)))).)))))).............. (-31.80 = -31.30 + -0.50)
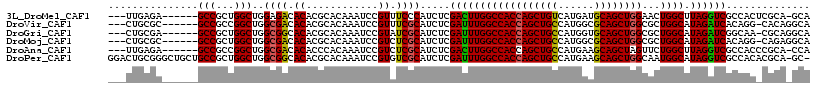
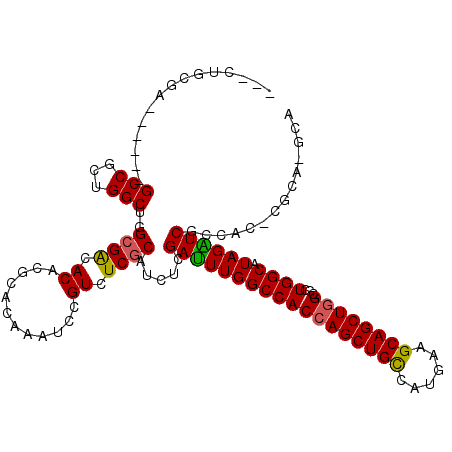
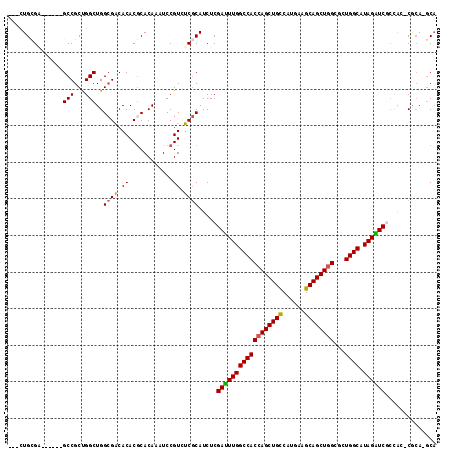
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:46 2006