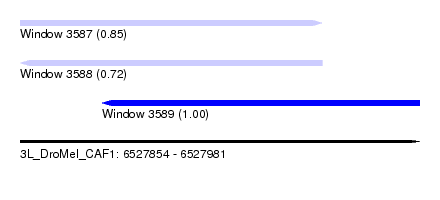
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,527,854 – 6,527,981 |
| Length | 127 |
| Max. P | 0.997082 |

| Location | 6,527,854 – 6,527,950 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.33 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6527854 96 + 23771897 AUUUCGGCUUUUUUGCAUUGUGUGUUGCCAUUUGUUUUGCUAUCGUCCUGUUUCCUGGGGCGUGGCACACAUACACAAAAAUAUA----UGUGGUUGUGU .....(((......((((...)))).))).........((((.((((((.......))))))))))(((((..((((........----))))..))))) ( -26.40) >DroSec_CAF1 19144 96 + 1 AUUUCGGCUUUUUUGCAUUGUGUGUUGCCAUUUGUUUUGCUAUCGUCCUGUUUCCUGGGGCGUGGCACACACACAAAUAUAUAUG----UGUGUGUGUGU .....(((......((((...)))).))).........((((.((((((.......))))))))))(((((((((........))----))))))).... ( -30.10) >DroEre_CAF1 19239 88 + 1 AUUUCGGCUUUUUUGCAUUGU--GUUGCCAUUUGUUUUGCUAUAGUCCUGUUUCCUGGGGCGUGGCACACACACAAA----UAUACGUGUCUGU------ ......((......))...((--((.(((((.(((......)))(((((.......))))))))))))))((((...----.....))))....------ ( -20.40) >consensus AUUUCGGCUUUUUUGCAUUGUGUGUUGCCAUUUGUUUUGCUAUCGUCCUGUUUCCUGGGGCGUGGCACACACACAAA_A_AUAUA____UGUGU_UGUGU ......((......)).((((((((((((((..(.....)....(((((.......))))))))))).))))))))........................ (-20.44 = -21.33 + 0.89)


| Location | 6,527,854 – 6,527,950 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -20.07 |
| Consensus MFE | -12.73 |
| Energy contribution | -13.73 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6527854 96 - 23771897 ACACAACCACA----UAUAUUUUUGUGUAUGUGUGCCACGCCCCAGGAAACAGGACGAUAGCAAAACAAAUGGCAACACACAAUGCAAAAAAGCCGAAAU ...........----....(((((((((.(((((((((((.((..(....).)).))...(.....)...)))).)))))..)))))))))......... ( -19.70) >DroSec_CAF1 19144 96 - 1 ACACACACACA----CAUAUAUAUUUGUGUGUGUGCCACGCCCCAGGAAACAGGACGAUAGCAAAACAAAUGGCAACACACAAUGCAAAAAAGCCGAAAU ...((((((((----((........))))))))))...((.((..(....).)).))...(((.......((....)).....))).............. ( -24.00) >DroEre_CAF1 19239 88 - 1 ------ACAGACACGUAUA----UUUGUGUGUGUGCCACGCCCCAGGAAACAGGACUAUAGCAAAACAAAUGGCAAC--ACAAUGCAAAAAAGCCGAAAU ------.............----(((((((((((((((....((.(....).))......(.....)...)))).))--)).)))))))........... ( -16.50) >consensus ACACA_ACACA____UAUAU_U_UUUGUGUGUGUGCCACGCCCCAGGAAACAGGACGAUAGCAAAACAAAUGGCAACACACAAUGCAAAAAAGCCGAAAU ...........................(((((((((((....((.(....).))......(.....)...)))).)))))))..((......))...... (-12.73 = -13.73 + 1.00)

| Location | 6,527,880 – 6,527,981 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.03 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -21.60 |
| Energy contribution | -22.16 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6527880 101 - 23771897 UUUCAGCUGUCAGUGUUCCACAUAUACAGGAACACAACCACA----UAUAUUUUUGUGUAUGUGUGCCACGCCCCAGGAAACAGGACGAUAGCAAAACAAAUGGC .....((((((.(((((((.........)))))))...((((----(((((....))))))))).........((.(....).))..))))))............ ( -31.40) >DroSec_CAF1 19170 101 - 1 UUCCAGCUACCAGUGUUUCACAUAUACACGAACACACACACA----CAUAUAUAUUUGUGUGUGUGCCACGCCCCAGGAAACAGGACGAUAGCAAAACAAAUGGC .....((((...((((.........)))).....((((((((----((........))))))))))...((.((..(....).)).)).))))............ ( -27.10) >DroEre_CAF1 19263 95 - 1 UUCCAGCUGUCACUGUUCCACAUAUACACAC------ACAGACACGUAUA----UUUGUGUGUGUGCCACGCCCCAGGAAACAGGACUAUAGCAAAACAAAUGGC .....((((((.(((((((......((((((------(((((........----)))))))))))((...))....)).))))))))...)))............ ( -26.80) >consensus UUCCAGCUGUCAGUGUUCCACAUAUACACGAACACA_ACACA____UAUAU_U_UUUGUGUGUGUGCCACGCCCCAGGAAACAGGACGAUAGCAAAACAAAUGGC .....((((((.((((..((((((((((............................))))))))))..)))).((.(....).))..))))))............ (-21.60 = -22.16 + 0.56)

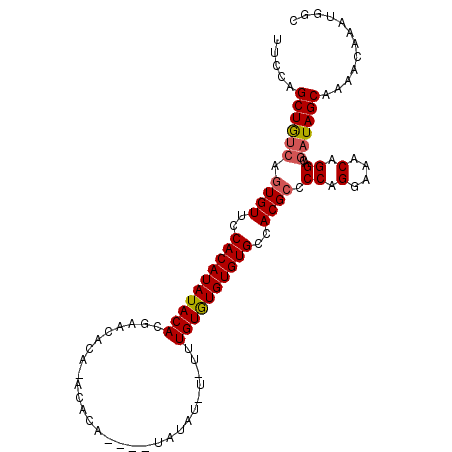
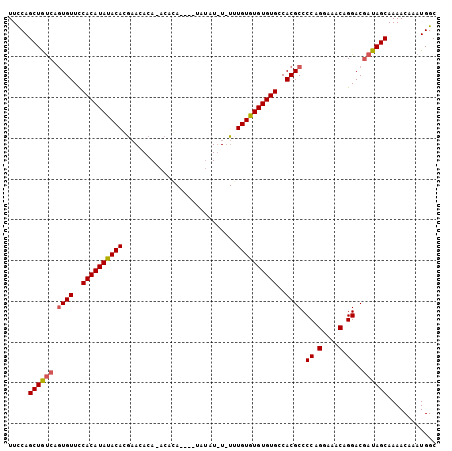
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:34 2006