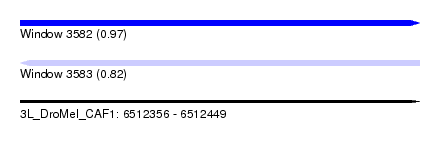
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,512,356 – 6,512,449 |
| Length | 93 |
| Max. P | 0.970216 |

| Location | 6,512,356 – 6,512,449 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.96 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.970216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
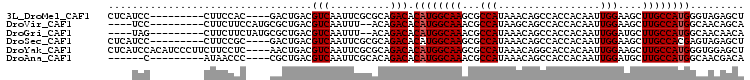
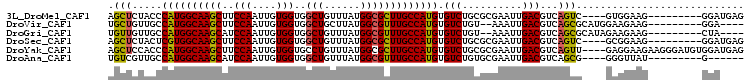
>3L_DroMel_CAF1 6512356 93 + 23771897 CUCAUCC---------CUUCCAC----GACUGACGUCAAUUCGCGCAGACACAUGGCAAGCGCCAUAAACAGCCACCACAAUUGGAAGCUUGCCAUGGGUAGAGCU (((..((---------(......----..(((.(((......))))))....(((((((((((........))..(((....)))..))))))))))))..))).. ( -29.30) >DroVir_CAF1 3622 91 + 1 ----UCC---------CUUCUUCCAUGCGCUGACGUCAAUUU--ACAGACACAUGGCAAACGCCAUAAGCAGCCACCACAAUUGGAAGCUUGCCAUGGCAACAGCA ----...---------............((((..(((.....--...))).((((((((..((.....)).((..(((....)))..))))))))))....)))). ( -26.00) >DroGri_CAF1 3832 91 + 1 ----UAG---------CUUCUUCUAUGCGCUGACGUCAAUUU--ACAGACACAUGGCAAACGCCAUAAACAGCCACCACAAUUGGAUGCUUGCCAUGGCAACAACA ----.((---------(........((.((((..(((.....--...)))..(((((....)))))...))))))(((....)))..)))(((....)))...... ( -22.10) >DroSec_CAF1 3896 93 + 1 CUCAUCC---------CUUCCGC----GACUGACGUCAAUUCGCGCAGACACAUGGCAAGCGCCAUAAACAGCCACCACAAUUGGAAGCUUGCCACGAGUAGAGCU (((....---------((..(((----((.((....))..))))).))((.(.((((((((((........))..(((....)))..)))))))).).)).))).. ( -25.90) >DroYak_CAF1 4084 102 + 1 CUCAUCCACAUCCCUUCUUCCUC----AACUGACGUCAAUUCGCGCAGACACAUGGCAAGCGCCAUAAACAGGCACCACAAUUGGAAGCUUGCCAUGGGUGGAGCU ....(((((..............----..(((.(((......))))))...(((((((((((((.......))).(((....)))..)))))))))).)))))... ( -38.20) >DroAna_CAF1 4509 87 + 1 ------C---------AUAACCC----CGCUGACGUCAAUUCGCACAGACACAUGGCAAACGCCAUAAACAGCCACCACAAUUGGAUGCUUGCCAUGGCAACGACA ------.---------.......----.((((..(((..........)))..(((((....)))))...))))..(((....))).((.((((....))))))... ( -19.30) >consensus ____UCC_________CUUCCUC____CACUGACGUCAAUUCGCACAGACACAUGGCAAACGCCAUAAACAGCCACCACAAUUGGAAGCUUGCCAUGGCAACAGCA ..................................(((..........))).((((((((...(((.................)))....))))))))......... (-14.80 = -14.96 + 0.17)
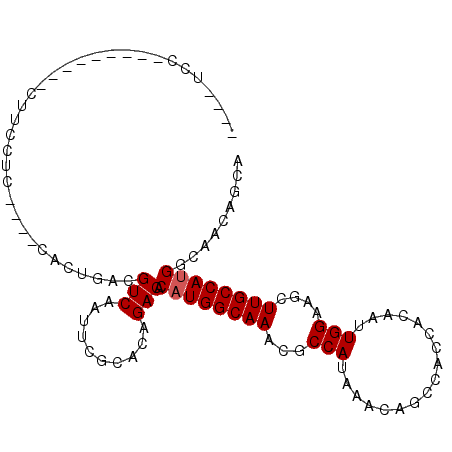
| Location | 6,512,356 – 6,512,449 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
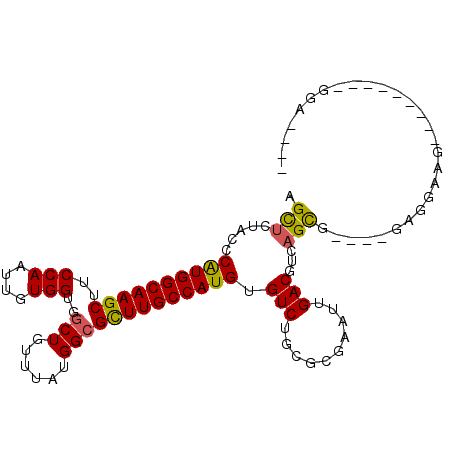
>3L_DroMel_CAF1 6512356 93 - 23771897 AGCUCUACCCAUGGCAAGCUUCCAAUUGUGGUGGCUGUUUAUGGCGCUUGCCAUGUGUCUGCGCGAAUUGACGUCAGUC----GUGGAAG---------GGAUGAG ..(((..((((((((((((..(((....)))..(((......)))))))))))).......(((((.(((....)))))----)))...)---------))..))) ( -33.30) >DroVir_CAF1 3622 91 - 1 UGCUGUUGCCAUGGCAAGCUUCCAAUUGUGGUGGCUGCUUAUGGCGUUUGCCAUGUGUCUGU--AAAUUGACGUCAGCGCAUGGAAGAAG---------GGA---- ..((.(((((((((((.(((.(((....))).)))))).)))))).....(((((((.(((.--..........))))))))))...)).---------)).---- ( -32.40) >DroGri_CAF1 3832 91 - 1 UGUUGUUGCCAUGGCAAGCAUCCAAUUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCUGU--AAAUUGACGUCAGCGCAUAGAAGAAG---------CUA---- ......(((....)))(((.((....((((.((((....((((((....)))))).(((...--.....))))))).)))).....)).)---------)).---- ( -27.10) >DroSec_CAF1 3896 93 - 1 AGCUCUACUCGUGGCAAGCUUCCAAUUGUGGUGGCUGUUUAUGGCGCUUGCCAUGUGUCUGCGCGAAUUGACGUCAGUC----GCGGAAG---------GGAUGAG ..((((((.((((((((((..(((....)))..(((......))))))))))))).))...(((((.(((....)))))----)))..))---------))..... ( -34.40) >DroYak_CAF1 4084 102 - 1 AGCUCCACCCAUGGCAAGCUUCCAAUUGUGGUGCCUGUUUAUGGCGCUUGCCAUGUGUCUGCGCGAAUUGACGUCAGUU----GAGGAAGAAGGGAUGUGGAUGAG ...(((((.((((((((((..(((....))).(((.......))))))))))))).((((...(...(..((....)).----.)....)...))))))))).... ( -35.20) >DroAna_CAF1 4509 87 - 1 UGUCGUUGCCAUGGCAAGCAUCCAAUUGUGGUGGCUGUUUAUGGCGUUUGCCAUGUGUCUGUGCGAAUUGACGUCAGCG----GGGUUAU---------G------ ..(((((((((((((((((..(((....)))..(((......))))))))))))).(((.((....)).)))).)))))----)......---------.------ ( -27.80) >consensus AGCUCUACCCAUGGCAAGCUUCCAAUUGUGGUGGCUGUUUAUGGCGCUUGCCAUGUGUCUGCGCGAAUUGACGUCAGCG____GAGGAAG_________GGA____ .(((.....((((((((((..(((....)))..(((......))))))))))))).(((..........)))...)))............................ (-22.24 = -21.72 + -0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:29 2006