
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 635,866 – 635,966 |
| Length | 100 |
| Max. P | 0.999880 |

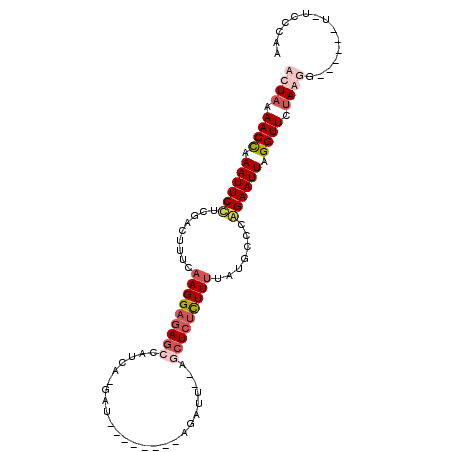
| Location | 635,866 – 635,966 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 64.83 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -11.10 |
| Energy contribution | -13.48 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
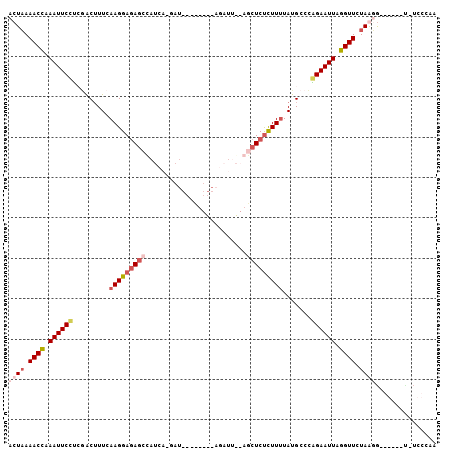
>3L_DroMel_CAF1 635866 100 + 23771897 ACUAAAACUAAAUUCCCCGACUUUCAAGGAGAGCUAUAAUUAUUAGCUUUAAGAUUGAAGCUCUCUUUUAUGCCCGGAAUUAGGUUCUAGUG------UUUCCCAA ((((.((((.((((((.........((((((((((.(((((...........))))).)))))))))).......)))))).)))).)))).------........ ( -26.09) >DroSec_CAF1 3202 83 + 1 ACUAAAACCAAAUUCCUAGACUUUCAAGGAGAGCUAUC-------------AGAUU--AGCUCUCUUCUAUGCCCGGAAUUUGGUUCUAGUG--------UCCCAA ((((.(((((((((((..(.(.....((((((((((..-------------....)--)))))))))....))..))))))))))).)))).--------...... ( -31.80) >DroEre_CAF1 6010 104 + 1 GCUAAAACCAAAUUCAUCAGCAUUAAAGGAGAG-CAUCA-GAUGCAUUUCAAGUUCGAAGCUCUUUUUUAUGCCUAGAAUUAGGUUCUAAGGUGGUAGCAUCCCAG ((((..(((..........((((.(((((((((-(.((.-.((.........))..)).)))))))))))))).((((((...)))))).)))..))))....... ( -29.90) >DroYak_CAF1 3401 87 + 1 GCUUAAACCAAAUUCUUGCACUUUUAAGGAUAGCCAUCA-GAU------------------UCCCUUUUAUGCAUAGAAUUAGGUUCUAAGAUGGUAGUGGCCCAA .((((((((.((((((((((.....((((..........-...------------------..))))...)))).)))))).)))).))))..(((....)))... ( -18.56) >consensus ACUAAAACCAAAUUCCUCGACUUUCAAGGAGAGCCAUCA_GAU________AGAUU__AGCUCUCUUUUAUGCCCAGAAUUAGGUUCUAAGG______U_UCCCAA ((((.((((.((((((.........(((((((((.........................))))))))).......)))))).)))).))))............... (-11.10 = -13.48 + 2.37)

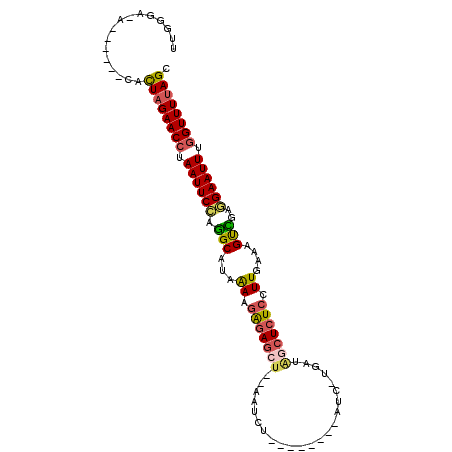
| Location | 635,866 – 635,966 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 64.83 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.54 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
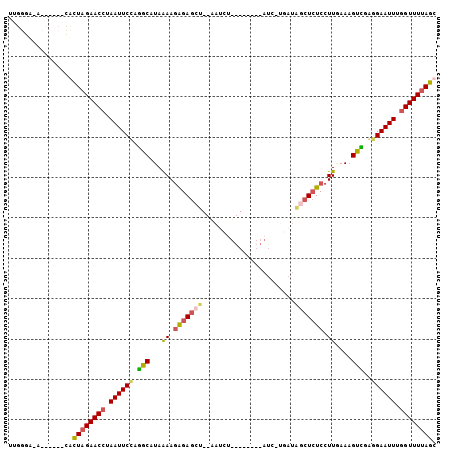
>3L_DroMel_CAF1 635866 100 - 23771897 UUGGGAAA------CACUAGAACCUAAUUCCGGGCAUAAAAGAGAGCUUCAAUCUUAAAGCUAAUAAUUAUAGCUCUCCUUGAAAGUCGGGGAAUUUAGUUUUAGU ...(....------)((((((((..((((((.(((...((.(((((((..(((.............)))..))))))).))....)))..))))))..)))))))) ( -26.72) >DroSec_CAF1 3202 83 - 1 UUGGGA--------CACUAGAACCAAAUUCCGGGCAUAGAAGAGAGCU--AAUCU-------------GAUAGCUCUCCUUGAAAGUCUAGGAAUUUGGUUUUAGU ......--------.((((((((((((((((((((...((.(((((((--(....-------------..)))))))).))....)))).)))))))))))))))) ( -37.60) >DroEre_CAF1 6010 104 - 1 CUGGGAUGCUACCACCUUAGAACCUAAUUCUAGGCAUAAAAAAGAGCUUCGAACUUGAAAUGCAUC-UGAUG-CUCUCCUUUAAUGCUGAUGAAUUUGGUUUUAGC .(((.......)))..((((((((.(((((..((((((((..(((((.(((....((.....))..-))).)-))))..))).)))))...))))).)))))))). ( -27.90) >DroYak_CAF1 3401 87 - 1 UUGGGCCACUACCAUCUUAGAACCUAAUUCUAUGCAUAAAAGGGA------------------AUC-UGAUGGCUAUCCUUAAAAGUGCAAGAAUUUGGUUUAAGC .(((.......))).((((((.((.((((((.(((((..((((((------------------..(-....)..).)))))....))))))))))).)))))))). ( -21.10) >consensus UUGGGA_A______CACUAGAACCUAAUUCCAGGCAUAAAAGAGAGCU__AAUCU________AUC_UGAUAGCUCUCCUUGAAAGUCGAGGAAUUUGGUUUUAGC ................((((((((.((((((.(((...((.(((((((.......................))))))).))....)))..)))))).)))))))). (-15.19 = -15.75 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:58 2006