
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,509,313 – 6,509,446 |
| Length | 133 |
| Max. P | 0.835366 |

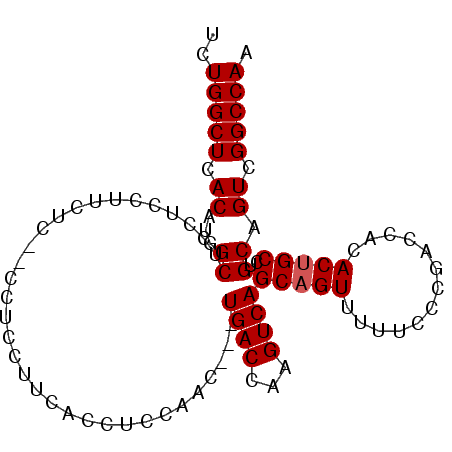
| Location | 6,509,313 – 6,509,406 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -19.93 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6509313 93 + 23771897 UCUGGCUCACAUGGCUCCUCCUUCUCUUCCUCCUUCACCUCCAAC---UGACCAAGUCAGCAGUUUUUCCCGACCACACCGCCUGCAGUCGGCCAA ..(((((.((..(((.............................(---((((...)))))..(((......)))......)))....)).))))). ( -16.30) >DroSec_CAF1 860 93 + 1 UCUGGCUCACAUGGCUCCUCCUUCUCUUCCGCCUUCACCUCCAAC---UGACCAAGUCAGCAGUUUUUCCCGACCACACUGCCUGCAGUCGGCCAA ..(((((.((..(((...............)))............---.......((..(((((.............)))))..)).)).))))). ( -17.38) >DroSim_CAF1 857 90 + 1 UCUGGCUCACAUGGCUUCUCCUCCUC---CUCCUUUACCUCCAAC---UGACCAAGUCAGCAGUUUUUCCCGACCACACUGCCUGCAGUCGGCCAA ..(((((.((..(((...........---...............(---((((...)))))..(((......)))......)))....)).))))). ( -16.00) >DroEre_CAF1 866 87 + 1 UCUGGCUCACAUGGCUUCG---------CCUGCUUCACCUCCAACUGCUGACCAAGUCAGCAGUUUUUCCCAAGCACACUGCCUGCAGUUGGCCAC ..(((((.((..(((....---------..(((((.......((((((((((...))))))))))......)))))....)))....)).))))). ( -30.02) >consensus UCUGGCUCACAUGGCUCCUCCUUCUC__CCUCCUUCACCUCCAAC___UGACCAAGUCAGCAGUUUUUCCCGACCACACUGCCUGCAGUCGGCCAA ..(((((.((...((.................................((((...))))(((((.............)))))..)).)).))))). (-14.27 = -14.52 + 0.25)

| Location | 6,509,313 – 6,509,406 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.90 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.53 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6509313 93 - 23771897 UUGGCCGACUGCAGGCGGUGUGGUCGGGAAAAACUGCUGACUUGGUCA---GUUGGAGGUGAAGGAGGAAGAGAAGGAGGAGCCAUGUGAGCCAGA (((((((((..((.....))..))))((.....((.((..(((..(((---.(....).)))..)))..))...))......))......))))). ( -26.20) >DroSec_CAF1 860 93 - 1 UUGGCCGACUGCAGGCAGUGUGGUCGGGAAAAACUGCUGACUUGGUCA---GUUGGAGGUGAAGGCGGAAGAGAAGGAGGAGCCAUGUGAGCCAGA (((((((((..((.....))..)))).......(.((((((...))))---)).)........(((...............)))......))))). ( -25.56) >DroSim_CAF1 857 90 - 1 UUGGCCGACUGCAGGCAGUGUGGUCGGGAAAAACUGCUGACUUGGUCA---GUUGGAGGUAAAGGAG---GAGGAGGAGAAGCCAUGUGAGCCAGA (((((((((..((.....))..)))).......(.((((((...))))---)).)..(((.......---...........)))......))))). ( -22.77) >DroEre_CAF1 866 87 - 1 GUGGCCAACUGCAGGCAGUGUGCUUGGGAAAAACUGCUGACUUGGUCAGCAGUUGGAGGUGAAGCAGG---------CGAAGCCAUGUGAGCCAGA .((((..((....(((.((.(((((.(....((((((((((...)))))))))).....).))))).)---------)...)))..))..)))).. ( -35.80) >consensus UUGGCCGACUGCAGGCAGUGUGGUCGGGAAAAACUGCUGACUUGGUCA___GUUGGAGGUGAAGGAGG__GAGAAGGAGAAGCCAUGUGAGCCAGA ....(((((((((.....)))))))))......(((((.((.((((...................................)))).)).)).))). (-20.65 = -20.53 + -0.12)

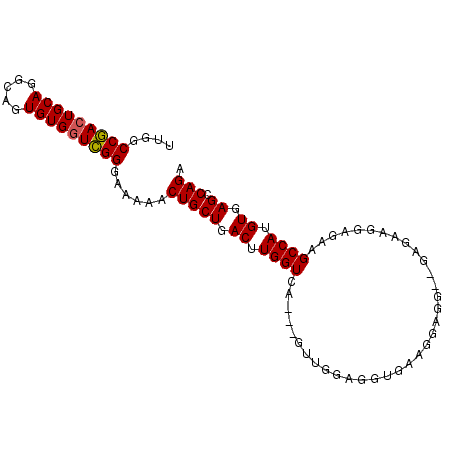

| Location | 6,509,338 – 6,509,446 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.92 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -22.14 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6509338 108 + 23771897 CUUCCUCCUUCACCUCCAAC---UGACCAAGUCAGCAGUUUUUCCCGACCACACCGCCUGCAGUCGGCCAACAAAGUGCUGAACUUCAAGUAGGAGUUGAGUCCCUUGGGG ....((((((((.((((.((---(....(((((((((.(((...(((((..((.....))..))))).....))).))))).))))..))).)))).))))......)))) ( -31.70) >DroSec_CAF1 885 108 + 1 CUUCCGCCUUCACCUCCAAC---UGACCAAGUCAGCAGUUUUUCCCGACCACACUGCCUGCAGUCGGCCAACAAACUGCUGAACUUCAAGGAGUAGUUGAGUCCCUUGGGA ..............(((((.---.((((((.((((((((((...(((((..((.....))..))))).....))))))))))((((....))))..))).)))..))))). ( -32.30) >DroSim_CAF1 882 105 + 1 C---CUCCUUUACCUCCAAC---UGACCAAGUCAGCAGUUUUUCCCGACCACACUGCCUGCAGUCGGCCAACAAAGUGCUGAACUUCAAGGAGGAGUUGAGUCCCUUGGGG .---(((..((.(((((...---(((.....((((((.(((...(((((..((.....))..))))).....))).))))))...))).)))))))..))).((....)). ( -30.50) >DroEre_CAF1 885 108 + 1 ---CCUGCUUCACCUCCAACUGCUGACCAAGUCAGCAGUUUUUCCCAAGCACACUGCCUGCAGUUGGCCACCAAAGUGCUGAACUUCAAGGAGCAGUUGAGUCCCUUAGGU ---.(((((((......((((((((((...)))))))))).......(((((((((....))))(((...)))..))))).........)))))))............... ( -35.30) >consensus C__CCUCCUUCACCUCCAAC___UGACCAAGUCAGCAGUUUUUCCCGACCACACUGCCUGCAGUCGGCCAACAAAGUGCUGAACUUCAAGGAGGAGUUGAGUCCCUUGGGG ....(((((((.....((.....))...(((((((((.(((...(((((..((.....))..))))).....))).))))).))))...)))))))......((....)). (-22.14 = -22.32 + 0.19)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:24 2006