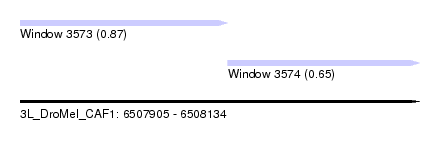
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,507,905 – 6,508,134 |
| Length | 229 |
| Max. P | 0.866298 |

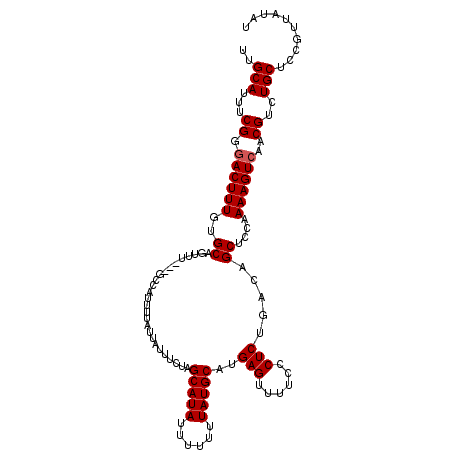
| Location | 6,507,905 – 6,508,024 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -24.28 |
| Energy contribution | -24.08 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6507905 119 + 23771897 AAAGCAAAUACAAUAAAUGAACGCCACCAGCGCUUCAUCGUCUUCUUCUUUUGGAGCGGAAGAACAGCUCUCCACUGGCAAGUUUGCCGCCUCUUGCUUAUCUACC-CUCCAACGAAAUG ................((((((((.....))).))))).......(((..((((((.((.(((..(((.......(((((....)))))......)))..))).))-)))))).)))... ( -27.42) >DroSec_CAF1 3126 119 + 1 AAAGCAAAUACAAUAAAUGAACGCCACCAGCGCUUCAUCGUCUUCUUCUCUUGGAGCGGAAGAACAGCUCUCCACUGGCAAGUUUGCCGCCUCUUGCUUAUCUACC-CUCCAACGAAAUG ................((((((((.....))).)))))..........((((((((.((.(((..(((.......(((((....)))))......)))..))).))-)))))).)).... ( -27.82) >DroSim_CAF1 3146 119 + 1 AAAGCAAAUACAAUAAAUGAACGCCACCAGCGCUUCAUCGUCUUCUUCGUUUGGAGCGGAAGAACAGCUCUCCACUGGCAAGUUUGCCUCCUCGUGCUUAUCUACC-CUCCAACGAAAUG ................((((((((.....))).))))).......((((((.((((.((.(((..(((........((((....)))).......)))..))).))-))))))))))... ( -31.06) >DroEre_CAF1 3122 119 + 1 AAAGCAAAUACAAUAAAUGAACGCCACCAGCGCUUUAUCGUCUUCUUCUUUUGGAGCGGAAGAACCGCUCUCCACUGGCAAGUUUGCUGCCUCUUGCUUAUCUACC-CUCCAACAAAAUG .((((((.........((((((((.....))).)))))..............(((((((.....))))))).....((((.(....)))))..)))))).......-............. ( -25.00) >DroYak_CAF1 3154 120 + 1 AAAGCAAAUACAAUAAAUGAACGCCAACAGCGCUUCAUCGUCUUCGUCUUUUGGAGCGGAAGAACAGCUCUCCACUGGCAAGUUUGCUGCUUUUUGCUUAUCUACCCCUCCAACAAAAUG .(((((((..((.(((((....((((.....((((((.((....)).....))))))(((.((.....)))))..))))..))))).))...)))))))..................... ( -25.80) >consensus AAAGCAAAUACAAUAAAUGAACGCCACCAGCGCUUCAUCGUCUUCUUCUUUUGGAGCGGAAGAACAGCUCUCCACUGGCAAGUUUGCCGCCUCUUGCUUAUCUACC_CUCCAACGAAAUG ................((((((((.....))).)))))............((((((.((.(((..(((........((((....)))).......)))..))).)).))))))....... (-24.28 = -24.08 + -0.20)

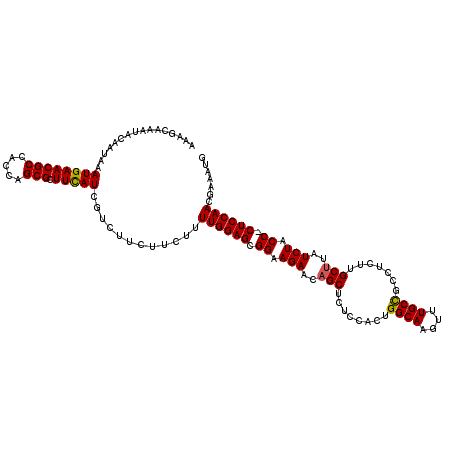
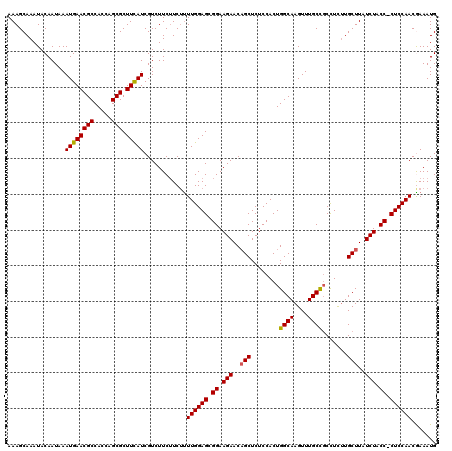
| Location | 6,508,024 – 6,508,134 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.41 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -17.76 |
| Energy contribution | -17.96 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6508024 110 + 23771897 UUGCAAUUCGGGACUUUGUGCAUUUU---GCCAUUUUAUUAUUUCUAGCAUAUUUUUUUAUGCAUGAGUUUUCCCUCUGACAGCUCCAAAAGUCAACGUCUGCUCCGUUAUAU ..(((...((.(((((((.((.....---)))...............(((((......)))))..(((((.((.....)).)))))..))))))..))..))).......... ( -19.70) >DroSec_CAF1 3245 110 + 1 UUGCAUUUCGGGACUUUGUGCAGUUU---GCCAUUUUAUUAUUUCUAGCAUAUUUUUUUAUGCAUGAGUUUUCCCUCUGACAGCUUCAAAAGUCAACGUCUGCUCCGUUGUAU .((((...(((((((((..((.(((.---..................(((((......)))))..(((......))).))).))....))))))...(....).))).)))). ( -20.60) >DroSim_CAF1 3265 110 + 1 UUGCAUUUCGGGACUUUGUGCAGUUU---GCCAUUUUAUUAUUUCUAGCAUAUUUUUUUAUGCAUGAGUUUUCCCUCUGACAGCUUCAAAAGUCAACGUCUGCUCCGUUAUAU ..(((...((.((((((..((.(((.---..................(((((......)))))..(((......))).))).))....))))))..))..))).......... ( -19.50) >DroEre_CAF1 3241 110 + 1 UUGCAUUUCGGGACUUUGUGCAGUUU---GGCAUUUUAUUAUUUCUAGCAUAUUUUUUUAUGCAUGAGUUUUCCCUCUGACAGCUCCAAAAGUCAACGUCUGCUCCUUUCUAU .........((((......((((.((---(((...............(((((......)))))..(((((.((.....)).))))).....)))))...))))))))...... ( -21.80) >DroYak_CAF1 3274 113 + 1 UUGCAUUUCGGUACUUUGUGCAGUUUUGUGCCAUUUUAUUAUUUCUAGCAUAUUUUUUUAUGCAUGAGUUUUCCCUCUGACAGCUCGAAAAGUCAACGUCUGCUCCUUUAUAU ..(((....(((((.............))))).........((((..(((((......)))))..(((((.((.....)).)))))))))..........))).......... ( -20.92) >consensus UUGCAUUUCGGGACUUUGUGCAGUUU___GCCAUUUUAUUAUUUCUAGCAUAUUUUUUUAUGCAUGAGUUUUCCCUCUGACAGCUCCAAAAGUCAACGUCUGCUCCGUUAUAU ..(((...((.((((((..((..........................(((((......)))))..(((......))).....))....))))))..))..))).......... (-17.76 = -17.96 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:21 2006