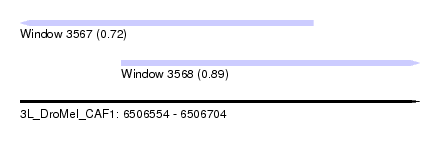
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,506,554 – 6,506,704 |
| Length | 150 |
| Max. P | 0.894056 |

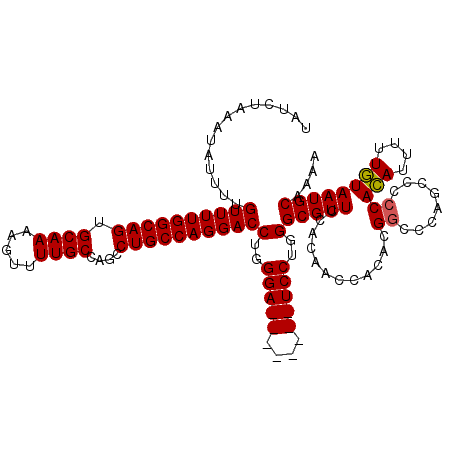
| Location | 6,506,554 – 6,506,664 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.27 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6506554 110 - 23771897 UAGAACGCCCAGGA--------UCCCAGGUCCUGGCAGGCUGGCAAAACUUUUGCACUGCCAAAACAAAAUAUUUAGAUAGCAAACGAACAUAUAUAUACACGCCAGGCUAA--ACACAC ......(((.((((--------(.....)))))))).(((..((((.....))))...))).................((((...((..............))....)))).--...... ( -23.94) >DroSec_CAF1 1876 110 - 1 UGGAACGCCCAGGA--------UCCCAGGUCCUGGCAGGCUGGCAAAAGUUUUGCACUGCCAAAACAAAAUAUUUAGAUAGCAAACGAACACAUAUAUACACGGCAGGCCAA--ACACAC (((...((((((((--------(.....))))))((.(((..((((.....))))...))).........(((....)))))....................)))...))).--...... ( -26.50) >DroSim_CAF1 1869 110 - 1 UGGAACGCCCAGGA--------UCCCAGGUCCUGGCAGGCUGGCAAAACUUUUGCACUGCCAAAACAAAAUAUUUAGAUAGCAAACGAACACAUAUAUACACGGCAGGCCAA--ACACAC (((...((((((((--------(.....))))))((.(((..((((.....))))...))).........(((....)))))....................)))...))).--...... ( -26.50) >DroEre_CAF1 1822 110 - 1 UGGAACGCCCAGGAUCCCAGGAUCCCAGGUCCUGGCAGGCUGGCAAAACUUUUGCACUGCCAAAACAAAAUAUUUAGAUAGCAAACGAACACAUACACAU----------AUAUACACAC .((...(((..(((((....)))))..)))))((((((....((((.....)))).))))))......................................----------.......... ( -25.40) >DroYak_CAF1 1794 112 - 1 UGGAACGCCCAGGA--------UCCCAGGUCCUGGCAGGCUGGCAAAACUUUUGCACUGCCAAAACAAAAUAUUUAGAUAGCAAACGAACACAUAUAUGCACGGCAGGCCAAACACACAC ......(((.((((--------(.....)))))))).(((..((((.....)))).(((((.........(((....)))(((..............)))..)))))))).......... ( -29.14) >consensus UGGAACGCCCAGGA________UCCCAGGUCCUGGCAGGCUGGCAAAACUUUUGCACUGCCAAAACAAAAUAUUUAGAUAGCAAACGAACACAUAUAUACACGGCAGGCCAA__ACACAC (((...(((..(((((....)))))..)))..((((((....((((.....)))).))))))..............................................)))......... (-19.34 = -19.90 + 0.56)

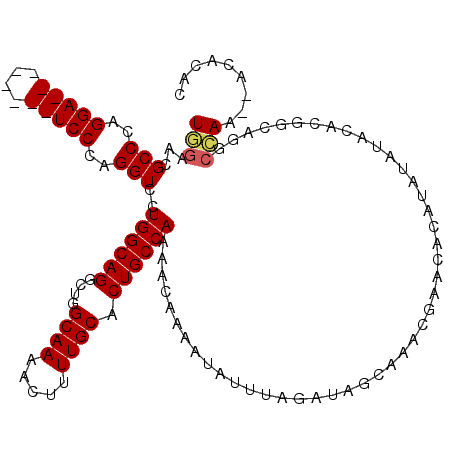
| Location | 6,506,592 – 6,506,704 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -26.94 |
| Energy contribution | -27.22 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6506592 112 + 23771897 UAUCUAAAUAUUUUGUUUUGGCAGUGCAAAAGUUUUGCCAGCCUGCCAGGACCUGGGA--------UCCUGGGCGUUCUACAACCACACGGGCCAGCCCCCACAUUUUUGUAAUGCAAAA ....................(((.(((((((((...((..((((((((((((....).--------))))))(.(((....))))...)))))..))......))))))))).))).... ( -29.20) >DroSec_CAF1 1914 112 + 1 UAUCUAAAUAUUUUGUUUUGGCAGUGCAAAACUUUUGCCAGCCUGCCAGGACCUGGGA--------UCCUGGGCGUUCCACAAGCACACGGCCCAGCCCCCAAAUUUUUUUAAUGCAAAA ..............((((((((((.((((.....))))....)))))))))).((((.--------..((((((................))))))..)))).................. ( -31.79) >DroSim_CAF1 1907 112 + 1 UAUCUAAAUAUUUUGUUUUGGCAGUGCAAAAGUUUUGCCAGCCUGCCAGGACCUGGGA--------UCCUGGGCGUUCCACAAGCACACGGCCCAGCCCCCACAUUUUUGUAAUGCAAAA ..............((((((((((.((((.....))))....)))))))))).((((.--------..((((((................))))))..))))...(((((.....))))) ( -32.59) >DroEre_CAF1 1852 120 + 1 UAUCUAAAUAUUUUGUUUUGGCAGUGCAAAAGUUUUGCCAGCCUGCCAGGACCUGGGAUCCUGGGAUCCUGGGCGUUCCACAACCACACGGCCCACCCCCCACAUUUUUGUAAUGCAAAA ..............((((((((((.((((.....))))....))))))))))(..(((((....)))))..)(((((..((((......((........))......))))))))).... ( -33.70) >DroYak_CAF1 1834 108 + 1 UAUCUAAAUAUUUUGUUUUGGCAGUGCAAAAGUUUUGCCAGCCUGCCAGGACCUGGGA--------UCCUGGGCGUUCCACAACCACACGGCCGAG----CACAUUUUUGUAAUGCAAAA ....................(((.(((((((((..((((.(((..(((((((....).--------))))))(.(((....))))....))).).)----)).))))))))).))).... ( -29.50) >consensus UAUCUAAAUAUUUUGUUUUGGCAGUGCAAAAGUUUUGCCAGCCUGCCAGGACCUGGGA________UCCUGGGCGUUCCACAACCACACGGCCCAGCCCCCACAUUUUUGUAAUGCAAAA ..............((((((((((.((((.....))))....))))))))))(..(((((....)))))..)(((((............((........))(((....)))))))).... (-26.94 = -27.22 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:15 2006