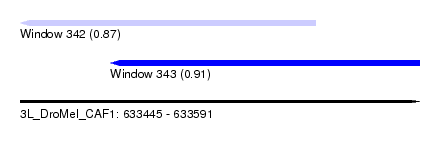
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 633,445 – 633,591 |
| Length | 146 |
| Max. P | 0.905425 |

| Location | 633,445 – 633,553 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -12.78 |
| Energy contribution | -13.94 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 633445 108 - 23771897 AAAAAAUUCCAC-CACAACAAAUACUGCUCA----AUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCAUUUAUGGUUGAAUAAAAAUAUGAAA-------AAGUAUCAGC ............-........(((((..(((----...(((((((..((.(((((((...(((.....)))..))))))).))..)))))))......)))..-------.))))).... ( -20.80) >DroSec_CAF1 752 117 - 1 AA--AAUUCCAC-CAUAACAAAUAUUGCUCAAUCAAUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCACUUAUGGUUGAAUAAAAAUAUGGAAAACAUUAAAGUAUCAGC ..--.((((.((-((((......((((((.(((......))).)))))).(((((((...(((.....)))..))))))))))))).))))............................. ( -25.10) >DroSim_CAF1 932 113 - 1 AA--AAUUCCAC-CACAACAAAUAUUACUCA----AUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCACUUAUGGUUGAAUAAAAAUAUGGAAAACAUUAAAGUAUCAGC ..--..(((((.-..........(((....)----)).(((((((..((.(((((((...(((.....)))..))))))).))..)))))))......)))))................. ( -21.80) >DroEre_CAF1 3621 101 - 1 AA--AAUUCCACAAAAAAC----AUUGCUCA----UCCUAUUCAGUAAUCAGUAGGCGUAAGCGCAGAGCUAUGAGCAUUUGGAGUUGAAUAAAAAUAUGAAAAACAUCAU--------- ..--(((((((.......(----((.((((.----..(((((.(....).)))))(((....))).)))).)))......)))))))..........((((......))))--------- ( -21.12) >DroYak_CAF1 936 114 - 1 AG--AAUUCAGUAGAAAACAAAUAUUGCUUA----UACUACUCAGUAAUCAGUACGCGUAAGCACAAAGCUAUGACCAUUUAGAGUUGAAUAGAAAUAUGAAAAACAUUCAAGUAUCAAC ..--.....................((((((----..(((((.(....).)))).)..))))))(((..(((........)))..)))....((.((.((((.....)))).)).))... ( -16.10) >consensus AA__AAUUCCAC_CAAAACAAAUAUUGCUCA____AUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCAUUUAUGGUUGAAUAAAAAUAUGAAAAACAUUAAAGUAUCAGC ......................................(((((((..((.((((((((((.((.....)))))))))))).))..)))))))............................ (-12.78 = -13.94 + 1.16)

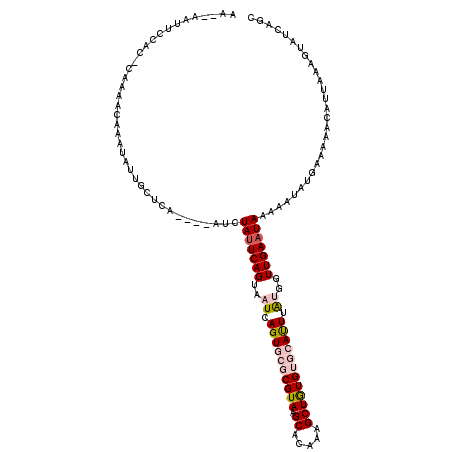
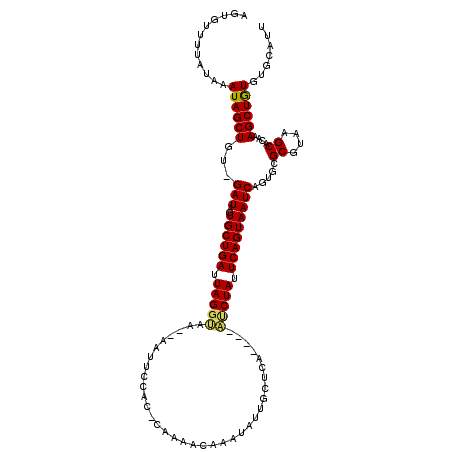
| Location | 633,478 – 633,591 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.44 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.09 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 633478 113 - 23771897 UGUGUUCUUUAAAUAGCUGUUGAUAGUGCUGAUUAGGUAAAAAAUUCCAC-CACAACAAAUACUGCUCA----AUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCAUU .((((.......((((((.(((...(..(((((..(((..........))-)........(((((....----.......))))))))))..)((....)).)))))))))..)))). ( -25.60) >DroSec_CAF1 792 114 - 1 AUUUUUUUAUAAAUAGCUGU-GAUAGUGCUGAUUAGGUAA--AAUUCCAC-CAUAACAAAUAUUGCUCAAUCAAUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCACU ...............((((.-(((((...(((((..((((--.(((....-.......))).))))..)))))..)))))))))....(((((((...(((.....)))..))))))) ( -26.90) >DroSim_CAF1 972 110 - 1 AGUUUUUUAUAAAUAGCUGU-GAUAGUGCUGAUUAGGUAA--AAUUCCAC-CACAACAAAUAUUACUCA----AUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCACU ............((((((.(-(...(..((((((((((..--......))-).................----..((....)))))))))..)((....)).)).))))))....... ( -24.10) >DroEre_CAF1 3652 107 - 1 AGCGAUUUAUAAAUAGCUGU-GAUAGUGCUGAUUAGGAAA--AAUUCCACAAAAAAC----AUUGCUCA----UCCUAUUCAGUAAUCAGUAGGCGUAAGCGCAGAGCUAUGAGCAUU .((.........((((((.(-(....((((((.(((((..--...............----........----))))).))))))........((....)).)).))))))..))... ( -25.41) >DroYak_CAF1 976 111 - 1 AGCGAUUUAUAAAUAGCUGU-GAUAGUGCUGAUUAGGUAG--AAUUCAGUAGAAAACAAAUAUUGCUUA----UACUACUCAGUAAUCAGUACGCGUAAGCACAAAGCUAUGACCAUU ............((((((.(-(...(((((((((((((((--.((.(((((.........)))))...)----).)))))...))))))))))((....)).)).))))))....... ( -30.30) >consensus AGUGUUUUAUAAAUAGCUGU_GAUAGUGCUGAUUAGGUAA__AAUUCCAC_CAAAACAAAUAUUGCUCA____AUCUAUUCAGUAAUCAGUGCGCGUAAGCACAAAGCUGUGUGCAUU ............((((((...(((..((((((.(((((...................................))))).))))))))).....((....))....))))))....... (-19.29 = -19.09 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:56 2006