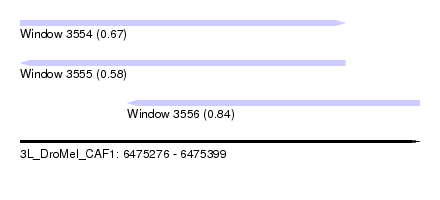
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,475,276 – 6,475,399 |
| Length | 123 |
| Max. P | 0.844729 |

| Location | 6,475,276 – 6,475,376 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -17.74 |
| Energy contribution | -17.92 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6475276 100 + 23771897 G-ACUCAGUGGCUCUC--UGUUUGUGUUUUAAA-AUUUUAAACGUUGCAAAUGUUUUGCAACAUUUCGACGGCUUUACCAGCUGUGUGCUACUAGUAUUCCACU .-(((.((((((.(..--.(((((.........-....)))))(((((((.....)))))))......((((((.....))))))).)))))))))........ ( -25.32) >DroSec_CAF1 9802 101 + 1 AAACUCAGUGGCUCUC--UGUUUGUGUUUUAAA-AUUUUAAACGUUGCAAAUGUUUUGAAACAUUUCGACGGCUUUACCAGCUGUGUGCUACUAGUAUUCCACG ..(((.((((((.(..--((...((((((((((-((................))))))))))))..))((((((.....))))))).)))))))))........ ( -24.19) >DroSim_CAF1 9896 100 + 1 A-ACUCAGUGGCUCUC--UGUUUGUGUUUUAAA-AUUUUAAACGUUGCAAAUGUUUUGCAACAUUUCGGCGGCUUUACCAGCUGUGUGCUACUAGUAUUCCACG .-(((.((((((.(..--.(((((.........-....)))))(((((((.....)))))))......((((((.....))))))).)))))))))........ ( -25.52) >DroEre_CAF1 10144 100 + 1 A-ACUCGCUGGUUCUC--UGUUUGUAUUUUGAA-AUUUUAAACGUUGCAAAUGUUUUGCAACAUUUCGACGGCUUUACCAGCUGUAUGCUACUAGUAUUCCUCU .-....((((((...(--((((.....(((((.-...))))).(((((((.....))))))).....)))))....))))))...((((.....))))...... ( -22.00) >DroYak_CAF1 10126 103 + 1 A-UUUCAGUGGUUCUCAGUGUUUGUAUUUUGAAAAUUUUAAACGUUGCAAAUGUUUUGCAACAUUUCGACGGCUUUACCAGCUGUGUGCUACUAGUAUUUCCCU .-....((((((.(.((((....(((.(((((.....)))))(((((.(((((((....))))))))))))....)))..)))).).))))))........... ( -22.20) >DroAna_CAF1 8888 83 + 1 A-CU--------GCAC--UGUUUGUAUUUUAAA-UUUUUAAGCGUUGCAAGUGUUUUGCAACAUUU-GACGGCCCUACCAGCUGUCCA--------AUCCCACU .-..--------(((.--((((((.........-....)))))).)))(((((((....)))))))-((((((.......))))))..--------........ ( -16.72) >consensus A_ACUCAGUGGCUCUC__UGUUUGUAUUUUAAA_AUUUUAAACGUUGCAAAUGUUUUGCAACAUUUCGACGGCUUUACCAGCUGUGUGCUACUAGUAUUCCACU ......((((((.......(((((..............)))))((((((((...))))))))......((((((.....))))))..))))))........... (-17.74 = -17.92 + 0.18)



| Location | 6,475,276 – 6,475,376 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -15.07 |
| Energy contribution | -16.57 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6475276 100 - 23771897 AGUGGAAUACUAGUAGCACACAGCUGGUAAAGCCGUCGAAAUGUUGCAAAACAUUUGCAACGUUUAAAAU-UUUAAAACACAAACA--GAGAGCCACUGAGU-C (((((..(((((((........))))))).........(((((((((((.....))))))))))).....-...............--.....)))))....-. ( -24.80) >DroSec_CAF1 9802 101 - 1 CGUGGAAUACUAGUAGCACACAGCUGGUAAAGCCGUCGAAAUGUUUCAAAACAUUUGCAACGUUUAAAAU-UUUAAAACACAAACA--GAGAGCCACUGAGUUU .((((..(((((((........)))))))....(((.((((((((....))))))).).)))........-...............--.....))))....... ( -18.30) >DroSim_CAF1 9896 100 - 1 CGUGGAAUACUAGUAGCACACAGCUGGUAAAGCCGCCGAAAUGUUGCAAAACAUUUGCAACGUUUAAAAU-UUUAAAACACAAACA--GAGAGCCACUGAGU-U .((((..(((((((........)))))))...))))..(((((((((((.....))))))))))).....-.............((--(.......)))...-. ( -25.00) >DroEre_CAF1 10144 100 - 1 AGAGGAAUACUAGUAGCAUACAGCUGGUAAAGCCGUCGAAAUGUUGCAAAACAUUUGCAACGUUUAAAAU-UUCAAAAUACAAACA--GAGAACCAGCGAGU-U ...((..(((((((........)))))))...)).((((((((((((((.....)))))))))))....(-(((............--)))).....)))..-. ( -22.60) >DroYak_CAF1 10126 103 - 1 AGGGAAAUACUAGUAGCACACAGCUGGUAAAGCCGUCGAAAUGUUGCAAAACAUUUGCAACGUUUAAAAUUUUCAAAAUACAAACACUGAGAACCACUGAAA-U ..((...(((((((........)))))))...)).((((((((((((((.....)))))))))))....((((((............))))))....)))..-. ( -22.50) >DroAna_CAF1 8888 83 - 1 AGUGGGAU--------UGGACAGCUGGUAGGGCCGUC-AAAUGUUGCAAAACACUUGCAACGCUUAAAAA-UUUAAAAUACAAACA--GUGC--------AG-U ....(.((--------(((((.(((.....))).)))-....(((((((.....))))))).........-.............))--)).)--------..-. ( -18.30) >consensus AGUGGAAUACUAGUAGCACACAGCUGGUAAAGCCGUCGAAAUGUUGCAAAACAUUUGCAACGUUUAAAAU_UUUAAAACACAAACA__GAGAGCCACUGAGU_U ...((..(((((((........)))))))...))....(((((((((((.....)))))))))))....................................... (-15.07 = -16.57 + 1.50)



| Location | 6,475,309 – 6,475,399 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -9.72 |
| Energy contribution | -10.88 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6475309 90 - 23771897 UACAACCUAUAUAUAUA----AAAAAAAGUGGAAUACUAGUAGCACACAGCU--GGUAAAGCCGUCGAAAUGUUGC---AAAACAUUUGCAACGUUUAA .................----.......((((..(((((((........)))--))))...))).).(((((((((---((.....))))))))))).. ( -21.00) >DroPse_CAF1 11877 81 - 1 GACAA-----AUAUA------A-AAAAAGUGGAAU------AGCACACAGCUGUGGCAAAGCCGUCAAAUUGUUGCAACAAAACAUUCGAAACGUUUUA (((..-----.....------.-..........((------(((.....)))))(((...))))))....((((.......)))).............. ( -11.70) >DroSec_CAF1 9836 89 - 1 UACAACCUAUAUA-----AAAAAAAAACGUGGAAUACUAGUAGCACACAGCU--GGUAAAGCCGUCGAAAUGUUUC---AAAACAUUUGCAACGUUUAA .............-----......((((((((..(((((((........)))--))))...))((..(((((((..---..))))))))).)))))).. ( -17.70) >DroSim_CAF1 9929 94 - 1 UACAACCUAUAUAUAAAAAAAAAAAAACGUGGAAUACUAGUAGCACACAGCU--GGUAAAGCCGCCGAAAUGUUGC---AAAACAUUUGCAACGUUUAA ............................((((..(((((((........)))--))))...))))..(((((((((---((.....))))))))))).. ( -24.00) >DroEre_CAF1 10177 83 - 1 UACAACCCAUAA-----------AAAAAGAGGAAUACUAGUAGCAUACAGCU--GGUAAAGCCGUCGAAAUGUUGC---AAAACAUUUGCAACGUUUAA ............-----------.....((((..(((((((........)))--))))...)).)).(((((((((---((.....))))))))))).. ( -21.60) >DroYak_CAF1 10162 88 - 1 UACAACAUAUAUAUA------AUAAAGAGGGAAAUACUAGUAGCACACAGCU--GGUAAAGCCGUCGAAAUGUUGC---AAAACAUUUGCAACGUUUAA ...............------.....((.((...(((((((........)))--))))...)).)).(((((((((---((.....))))))))))).. ( -24.60) >consensus UACAACCUAUAUAUA______AAAAAAAGUGGAAUACUAGUAGCACACAGCU__GGUAAAGCCGUCGAAAUGUUGC___AAAACAUUUGCAACGUUUAA ..............................((..((((...(((.....)))..))))...))....(((((((((............))))))))).. ( -9.72 = -10.88 + 1.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:03 2006