| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 630,139 – 630,237 |
| Length | 98 |
| Max. P | 0.652348 |

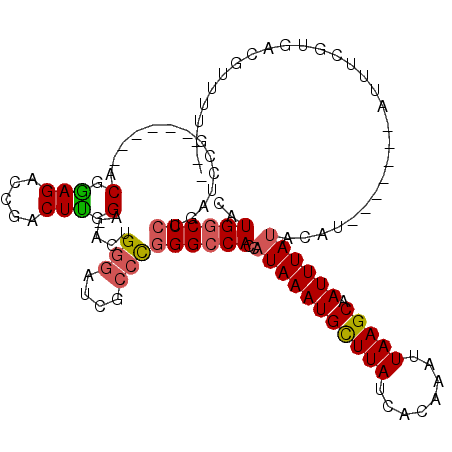
| Location | 630,139 – 630,237 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 630139 98 + 23771897 ---AAAACGUCACGAAAU-------AUGUAUAAAUUGCUUAAUUUGUGAUAAGCAUUUAUGGUGGCCCGGGCGAUCCCAGUUUCCAAGUCGGUCUCCGU------UG------GGCCAUG ---.....((((((((.(-------(.(((.....))).)).))))))))...........((((((((((....)))...........(((...))).------.)------)))))). ( -28.40) >DroSec_CAF1 41571 108 + 1 --GAAAACGUCACGAAAUACA---UAUGUAUAAAUUGCUUAAUUUGUGAUAAGCAUUUAUGGUGGCCCGGGCGAUCCCAGU-UCCAAGUCGGUCUCCGU------CGGAUCAGGGUCAUG --...................---....(((((((.(((((........))))))))))))((((((((((....))).(.-(((..(.(((...))).------)))).).))))))). ( -27.50) >DroSim_CAF1 39822 104 + 1 --GAAAACGUCACGAAAU-------AUGUAUAAAUUGCUUAAUUUGUGAUAAGCAUUUAUGGUGGCCCGGGCGAUCCCAGU-UCCAAGUCGGUCUCCGU------CGGAUCAGGGCCAUG --......((((((((.(-------(.(((.....))).)).))))))))...........((((((((((....))).(.-(((..(.(((...))).------)))).).))))))). ( -32.40) >DroYak_CAF1 46271 113 + 1 CGAAAAACGCCAUGAAAUAUGUACUAUGUAUAAAUUGCUUAAUUUGUGAUAAGCAUUUAUGGUGGCCCAGGCGAUCCCAGU-GCCAAGUCGCUCUUCGU------CGGCUCAGGGCCAUG ........((((((((...(((((...)))))...((((((........))))))))))))))(((((.((((.......)-))).((((((.....).------)))))..)))))... ( -34.80) >DroAna_CAF1 51582 103 + 1 ---AAAAUGCCACGAAAU-----------GUAAAUUGCUUAAUUUGUGAUAAACAUUUAUGGUGGCCCAGGCGG---CAGAGUCGGAGUCGGUCUCCGAGUCGAACGGAUCAGGGCCAUG ---.....((((..((((-----------((..((..(.......)..))..)))))).))))(((((.((((.---..((.((((((.....)))))).))...))..)).)))))... ( -36.70) >consensus ___AAAACGUCACGAAAU_______AUGUAUAAAUUGCUUAAUUUGUGAUAAGCAUUUAUGGUGGCCCGGGCGAUCCCAGU_UCCAAGUCGGUCUCCGU______CGGAUCAGGGCCAUG ............................(((((((.(((((........))))))))))))((((((((((((((............)))).)))(((.......)))....))))))). (-20.36 = -20.36 + -0.00)

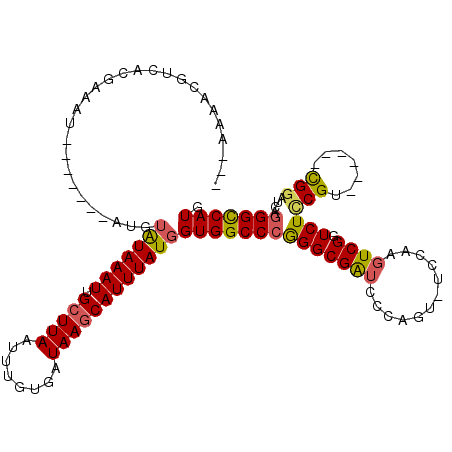
| Location | 630,139 – 630,237 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 630139 98 - 23771897 CAUGGCC------CA------ACGGAGACCGACUUGGAAACUGGGAUCGCCCGGGCCACCAUAAAUGCUUAUCACAAAUUAAGCAAUUUAUACAU-------AUUUCGUGACGUUUU--- ..(((((------(.------.(((...)))....(....).(((....)))))))))..(((((((((((........))))).))))))....-------...............--- ( -26.60) >DroSec_CAF1 41571 108 - 1 CAUGACCCUGAUCCG------ACGGAGACCGACUUGGA-ACUGGGAUCGCCCGGGCCACCAUAAAUGCUUAUCACAAAUUAAGCAAUUUAUACAUA---UGUAUUUCGUGACGUUUUC-- ((((((((...((((------((((...)))..)))))-...(((....)))))).....(((((((((((........))))).)))))).....---......)))))........-- ( -25.40) >DroSim_CAF1 39822 104 - 1 CAUGGCCCUGAUCCG------ACGGAGACCGACUUGGA-ACUGGGAUCGCCCGGGCCACCAUAAAUGCUUAUCACAAAUUAAGCAAUUUAUACAU-------AUUUCGUGACGUUUUC-- ..((((((...((((------((((...)))..)))))-...(((....)))))))))..(((((((((((........))))).))))))....-------................-- ( -29.60) >DroYak_CAF1 46271 113 - 1 CAUGGCCCUGAGCCG------ACGAAGAGCGACUUGGC-ACUGGGAUCGCCUGGGCCACCAUAAAUGCUUAUCACAAAUUAAGCAAUUUAUACAUAGUACAUAUUUCAUGGCGUUUUUCG ..((((.....))))------.(((((((((.((((((-.(..((....))..)))))..(((((((((((........))))).))))))..................))))))))))) ( -33.60) >DroAna_CAF1 51582 103 - 1 CAUGGCCCUGAUCCGUUCGACUCGGAGACCGACUCCGACUCUG---CCGCCUGGGCCACCAUAAAUGUUUAUCACAAAUUAAGCAAUUUAC-----------AUUUCGUGGCAUUUU--- ..((((((.....((...((.((((((.....)))))).))..---.))...))))))((((((((((..((..(.......)..))..))-----------)))).))))......--- ( -28.60) >consensus CAUGGCCCUGAUCCG______ACGGAGACCGACUUGGA_ACUGGGAUCGCCCGGGCCACCAUAAAUGCUUAUCACAAAUUAAGCAAUUUAUACAU_______AUUUCGUGACGUUUU___ ..((((((..............(.(((.....))).).....(((....)))))))))..(((((((((((........))))).))))))............................. (-19.00 = -19.28 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:54 2006