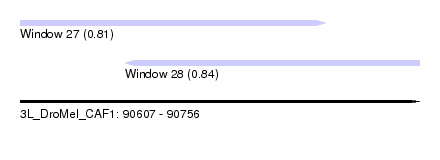
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 90,607 – 90,756 |
| Length | 149 |
| Max. P | 0.842543 |

| Location | 90,607 – 90,721 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -18.40 |
| Energy contribution | -19.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 90607 114 + 23771897 GGUUUGGUCAAUAAAACAAACAACAAAAAAUAAAAGUGUGCGAGUGGCUGUACGCAAAUCAGGGCAUAUAUGGAUACACA----UACAUAAAAACAUACAUAAGUAUGCUUGGAACAC .(((((..........)))))..............(((((((......)))))))...(((((.....((((......))----))........(((((....))))))))))..... ( -20.80) >DroSec_CAF1 35992 116 + 1 GGUUUGGUCAACAAAACAAACAACAAAAAAUGAAAGUGUGAGAGCGGCUGCUCGCAAACCAGGGCAUAUACAUAUGUACAUAUGUACAUAAGUACAU--AUAAGUAUGCUUGAAACAC ((((((.................((....((....)).)).((((....)))).)))))).((((((((..((((((((.(((....))).))))))--))..))))))))....... ( -30.90) >DroSim_CAF1 41157 116 + 1 GGUUUGGUCAACAAAACAAACAACAAAAAAUAAAAGUGUGAGAGCGGCUGUACGCAAACCAGGGCAUAUACAUAUGUACAUAUGUACAUAAGUACAU--AUAAGUAUGCUUGAAACAC .(((((..........)))))..............((((....(((......)))......((((((((..((((((((.(((....))).))))))--))..))))))))...)))) ( -28.80) >consensus GGUUUGGUCAACAAAACAAACAACAAAAAAUAAAAGUGUGAGAGCGGCUGUACGCAAACCAGGGCAUAUACAUAUGUACAUAUGUACAUAAGUACAU__AUAAGUAUGCUUGAAACAC .(((((..........)))))..............((((....(((......)))......((((((((((.(((((((....))))))).))..........))))))))...)))) (-18.40 = -19.40 + 1.00)

| Location | 90,646 – 90,756 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -30.61 |
| Consensus MFE | -24.10 |
| Energy contribution | -23.89 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 90646 110 - 23771897 GUUCUCGGAAAGUGCUCUGCAUUAAUUAUGGGAAUGUGUUCCAAGCAUACUUAUGUAUGUUUUUAUGUA----UGUGUAUCCAUAUAUGCCCUGAUUUGCGUACAGCCACUCGC ......((...((((...(((..(((((.(((..........((((((((....)))))))).((((((----((......)))))))))))))))))))))))..))...... ( -26.00) >DroSec_CAF1 36031 112 - 1 AUUCCCGCAAAGUGCUCUGCAUUAAUUAUGGGAAUGUGUUUCAAGCAUACUUAU--AUGUACUUAUGUACAUAUGUACAUAUGUAUAUGCCCUGGUUUGCGAGCAGCCGCUCUC (((((((.(((((((...)))))..)).)))))))(((..(((.(((((..(((--((((((.(((....))).)))))))))..)))))..)))..)))((((....)))).. ( -35.20) >DroSim_CAF1 41196 112 - 1 AUUCCCGCAAAGUGCUCUGCAUUAAUUAUGGGAAUGUGUUUCAAGCAUACUUAU--AUGUACUUAUGUACAUAUGUACAUAUGUAUAUGCCCUGGUUUGCGUACAGCCGCUCUC (((((((.(((((((...)))))..)).)))))))(((......(((.(((..(--((((((.(((((((....))))))).)))))))....))).))).......))).... ( -30.62) >consensus AUUCCCGCAAAGUGCUCUGCAUUAAUUAUGGGAAUGUGUUUCAAGCAUACUUAU__AUGUACUUAUGUACAUAUGUACAUAUGUAUAUGCCCUGGUUUGCGUACAGCCGCUCUC (((((((...(((((...))))).....)))))))(((......(((.(((.....((((((.(((((((....))))))).)))))).....))).))).......))).... (-24.10 = -23.89 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:07 2006