
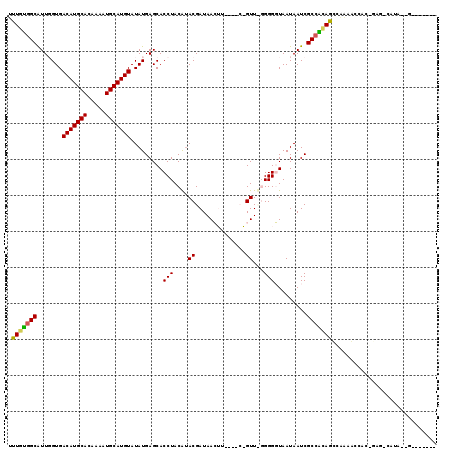
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,456,727 – 6,456,826 |
| Length | 99 |
| Max. P | 0.985042 |

| Location | 6,456,727 – 6,456,826 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.27 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.06 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
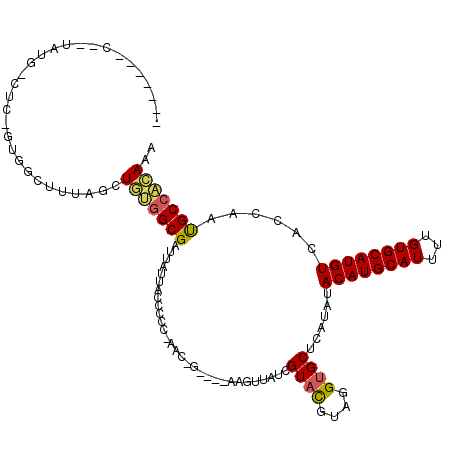
>3L_DroMel_CAF1 6456727 99 + 23771897 UUUGCCGCAUUGGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACGUACGAUAACUU----CGGUUCGGGGGUAAUAAUCGCCGCAGUCAAAACCAUUGC---------------- ......(((.((((.(((((((.....)))))))........(((..((..(((.....)----))...))..)))...................)))).)))---------------- ( -26.00) >DroVir_CAF1 269077 99 + 1 UCUCUGGCGUUUGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACAUACGAUAAG--------GUU--GGGUUAAUAAUCGCCAGAGA-G-CAACGG--AGCCACAAGUU------ (((((((((((..(((((((((.....))))))).))..)))(((((...((......--------)))--))))........))))))))-(-(.....--.))........------ ( -32.30) >DroPse_CAF1 248081 117 + 1 UUUGUGGCAUUGGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACAUACGAUAACUUUUUGCAGUUUGGGGGUAAUAAUCGCCACAGCUAAAGCCACAGAGUCAUA--GCCAGAGC ((((((((...(((((((((((.....))))))).......))))....................(((((((.(((....))).))).))))...))))))))......--........ ( -31.51) >DroGri_CAF1 200900 107 + 1 GCUCUGGCGUUUGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACGGACGAUAAG--------GUU--GGGUUAAUAAUCGCCACAGUAACAAGCAA--ACAGUUAGAGUUACACA ((((((((((((((.(((((((.....)))))))...((.((..(....).......(--------(((--(......))))))))).........))))--)).))))))))...... ( -28.90) >DroAna_CAF1 197623 101 + 1 UUUGUGGCAUUGGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACGUACGAUAACUC--CGCGGUUCGGGGGUAAUAAUCGCCGCAGCCAAAGCCACGGC---------------- .(((((((...(((((((((((.....))))))).......))))...............--...(((((((.(((....))).))).))))...))))))).---------------- ( -33.81) >DroPer_CAF1 255753 117 + 1 UUUGUGGCAUUGGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACAUACGAUAACUUUUUGCAGUUUGGGGGUAAUAAUCGCCACAGCUAAAGCCACAGAGUCAUA--GCCAGAGC ((((((((...(((((((((((.....))))))).......))))....................(((((((.(((....))).))).))))...))))))))......--........ ( -31.51) >consensus UUUGUGGCAUUGGUGACAUGCACAAAAUGCAUGUAUAUGAGCACCUACAUACGAUAACUU____C_GUU_GGGGGUAAUAAUCGCCACAGCCAAAACCAC_GAG_CAUA__G_______ .(((((((.......(((((((.....))))))).........(((....((..............))...))).........)))))))............................. (-20.53 = -20.06 + -0.47)


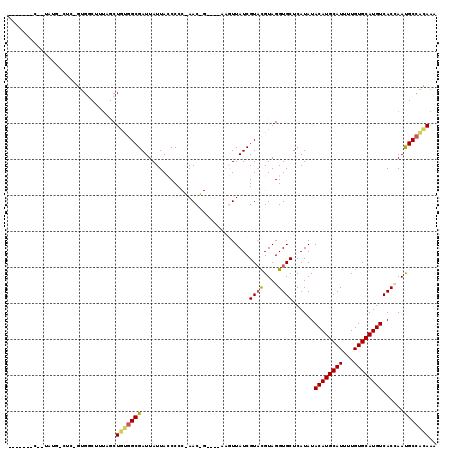
| Location | 6,456,727 – 6,456,826 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.27 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.08 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6456727 99 - 23771897 ----------------GCAAUGGUUUUGACUGCGGCGAUUAUUACCCCCGAACCG----AAGUUAUCGUACGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACCAAUGCGGCAAA ----------------(((.((((.....((((((((((.(((............----.))).))))).)))))..........((((((((...)))))))).)))).)))...... ( -26.22) >DroVir_CAF1 269077 99 - 1 ------AACUUGUGGCU--CCGUUG-C-UCUCUGGCGAUUAUUAACCC--AAC--------CUUAUCGUAUGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACAAACGCCAGAGA ------.....(..((.--..))..-)-(((((((((...........--...--------......(((((((.((....)).)))))))..((((((.....))))))))))))))) ( -29.50) >DroPse_CAF1 248081 117 - 1 GCUCUGGC--UAUGACUCUGUGGCUUUAGCUGUGGCGAUUAUUACCCCCAAACUGCAAAAAGUUAUCGUAUGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACCAAUGCCACAAA (((..(((--(((......))))))..)))(((((((.............((((......))))..........((((.......((((((((...))))))))))))..))))))).. ( -35.61) >DroGri_CAF1 200900 107 - 1 UGUGUAACUCUAACUGU--UUGCUUGUUACUGUGGCGAUUAUUAACCC--AAC--------CUUAUCGUCCGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACAAACGCCAGAGC .......((((...(((--(((...((..((((((((((.........--...--------...)))).))))))..))......((((((((...))))))))..))))))..)))). ( -24.06) >DroAna_CAF1 197623 101 - 1 ----------------GCCGUGGCUUUGGCUGCGGCGAUUAUUACCCCCGAACCGCG--GAGUUAUCGUACGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACCAAUGCCACAAA ----------------...(((((.((((((((((((((....((..(((.....))--).)).))))).)))))..........((((((((...))))))))..)))).)))))... ( -36.70) >DroPer_CAF1 255753 117 - 1 GCUCUGGC--UAUGACUCUGUGGCUUUAGCUGUGGCGAUUAUUACCCCCAAACUGCAAAAAGUUAUCGUAUGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACCAAUGCCACAAA (((..(((--(((......))))))..)))(((((((.............((((......))))..........((((.......((((((((...))))))))))))..))))))).. ( -35.61) >consensus _______C__UAUG_CUC_GUGGCUUUAGCUGUGGCGAUUAUUACCCCC_AAC_G____AAGUUAUCGUACGUAGGUGCUCAUAUACAUGCAUUUUGUGCAUGUCACCAAUGCCACAAA ..............................(((((((..............................((((....))))......((((((((...))))))))......))))))).. (-19.33 = -19.08 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:51 2006