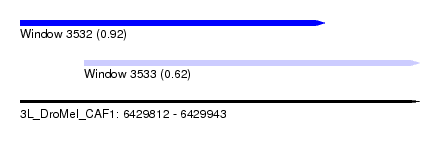
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,429,812 – 6,429,943 |
| Length | 131 |
| Max. P | 0.918244 |

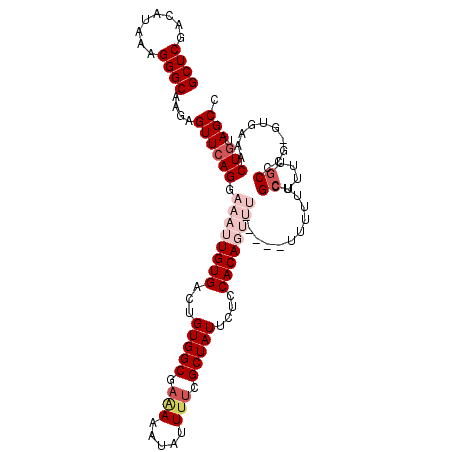
| Location | 6,429,812 – 6,429,912 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.98 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.28 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
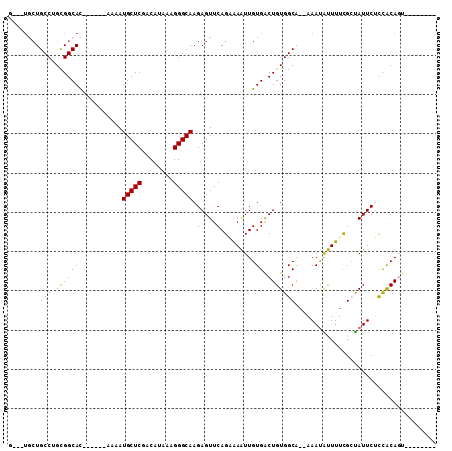
>3L_DroMel_CAF1 6429812 100 + 23771897 G---UGCUGCCAGCGGCAC------AAAAUGCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGCGAAAAAUAUUUUCGCUAUUCUCCACAGUUUUCCAUU (---(((((....))))))------....(((((........)))))........((((((((((...(((((((((.....)))))))))....)))))))))).... ( -36.50) >DroVir_CAF1 231499 89 + 1 -AGCAACUGCCUGUGGCAU------A-AAUGCUCGACAUAAAGGGCAAGAGUUCAGCGAGUUGUGACUGUGGCA---AAUGCUU-UGGUAUUCGCCACAGU-------- -.(((((((((...)))..------.-..(((((........)))))...........)))))).((((((((.---(((((..-..))))).))))))))-------- ( -33.20) >DroPse_CAF1 214550 96 + 1 G---UGCUGCCUGCGGCACAACGGCAAAAUGCUCGACAUAAAGGGCAAGAGUUCAGAAAAUUGUGACUGUGGCAAGAAAUAUUUUCGCUAUUCUCCACA---------- (---(((((....))))))...((.....(((((........)))))(((((...((((((.((..((......))..))))))))...)))))))...---------- ( -24.50) >DroMoj_CAF1 188143 89 + 1 -AAGUUGUGCCUGUGGCAC------A-AAUGCUCCACAGAAAGGGCAAGAGUUCAGCGAGUUGUGGCUGUGGCA---AACGCUU-CGGCAUUCGCUGCAGU-------- -...(((((((...)))))------)-).(((((........)))))......((((((((....((((.(((.---...))).-))))))))))))....-------- ( -31.90) >DroAna_CAF1 171203 93 + 1 G---UGCUGCCAGCGGCAC------AAAAUGCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGC--AAAAUAUUUUCGCUAUUCUUCACAGUUU-----U (---(((((....))))))------....(((((........))))).........((((((((((..(((((--...........)))))...)))))))))-----) ( -29.60) >DroPer_CAF1 221705 96 + 1 G---UGCUGCCUGCGGCACAACGGCAAAAUGCUCGACAUAAAGGGCAAGAGUUCAGAAAAUUGUGACUGUGGCAAGAAAUAUUUUCGCUAUUCUCCACA---------- (---(((((....))))))...((.....(((((........)))))(((((...((((((.((..((......))..))))))))...)))))))...---------- ( -24.50) >consensus G___UGCUGCCUGCGGCAC______AAAAUGCUCGACAUAAAGGGCAAGAGUUCAGAAAAUUGUGACUGUGGCA__AAAUAUUUUCGCUAUUCUCCACAGU________ .......((((.(((((((..........(((((........))))).(....)........))).))))))))................................... (-14.59 = -14.28 + -0.30)

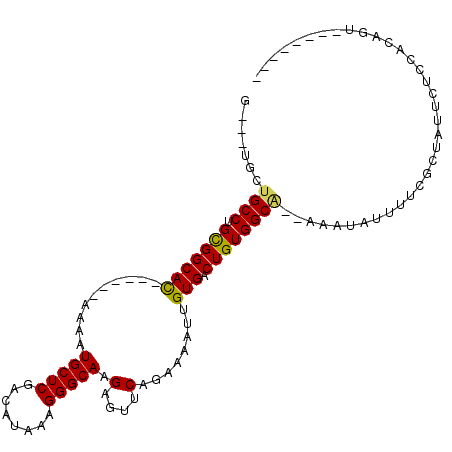
| Location | 6,429,833 – 6,429,943 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -31.19 |
| Consensus MFE | -12.98 |
| Energy contribution | -14.98 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6429833 110 + 23771897 GCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGCGAAAAAUAUUUUCGCUAUUCUCCACAGUUUUCCAUUUUUUUCGCUUUUGCCCG-GUGAAACUGUAGCC (((((.(((...((((((((((...((((((((((...(((((((((.....)))))))))....))))))))))..........)))))))))).-)))....)).))). ( -37.12) >DroPse_CAF1 214577 86 + 1 GCUCGACAUAAAGGGCAAGAGUUCAGAAAAUUGUGACUGUGGCAAGAAAUAUUUUCGCUAUUCUCCACA------------------------CAG-GAGAAACUGUAGCC ((((........))))....((((((......((((..(((........)))..))))..((((((...------------------------..)-))))).))).))). ( -20.40) >DroEre_CAF1 172146 109 + 1 GCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGCGAAAAAUAUU-UCGCUAUUCUUCACAGUUUUCCAUUUUUUUCGCUUUUGCCCG-GUGAAACUGUAGCC (((((.(((...((((((((((...(((((((((((..(((((((((....))-)))))))...)))))))))))..........)))))))))).-)))....)).))). ( -39.22) >DroYak_CAF1 181388 111 + 1 GCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGCGAAAAAUAUUUUCGCUAUUCUUCACAGUUUUCCAUUUUUUUCGCUUUUGCCCGGGUGAAACUGUAGCC (((((.(((...((((((((((...(((((((((((..(((((((((.....)))))))))...)))))))))))..........))))))))))..)))....)).))). ( -39.62) >DroAna_CAF1 171224 102 + 1 GCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGC--AAAAUAUUUUCGCUAUUCUUCACAGUUU-----UUGCUUCGCUUUUCC-CG-UCUGGUCUCUAGCC (((.(((.....(((.((((((...(((((((((((..(((((--...........)))))...)))))))))-----)).....))))))))-).-....)))...))). ( -30.40) >DroPer_CAF1 221732 86 + 1 GCUCGACAUAAAGGGCAAGAGUUCAGAAAAUUGUGACUGUGGCAAGAAAUAUUUUCGCUAUUCUCCACA------------------------CAG-GAGAAACUGUAGCC ((((........))))....((((((......((((..(((........)))..))))..((((((...------------------------..)-))))).))).))). ( -20.40) >consensus GCUCGACAUAAAGGGCAAGAGUUCAGGAAAUUGUGACUGUGGCGAAAAAUAUUUUCGCUAUUCUCCACAGUUU_____UUUUUUCGCUUUUGCCCG_GUGAAACUGUAGCC ((((........))))....((((((.((((((((...(((((.(((.....))).)))))....))))))))............((....))..........))).))). (-12.98 = -14.98 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:41 2006