
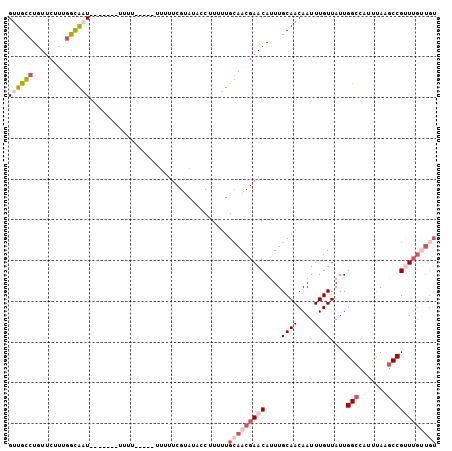
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,419,495 – 6,419,587 |
| Length | 92 |
| Max. P | 0.940885 |

| Location | 6,419,495 – 6,419,587 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -18.15 |
| Consensus MFE | -11.84 |
| Energy contribution | -13.53 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6419495 92 + 23771897 GUUGCCUGUUCUUUGGCAAU-------UUU------UUUUUCGUAUACCUUUUUGCAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUUAAGCCGUUUGUUGU ((((((........))))))-------...------..................(((((((((....((((....))))....(((.......)))))))))))) ( -21.10) >DroSec_CAF1 157504 93 + 1 GUUGCCUGUUCUUUGGCAAU-------UUUU-----UUUUUCGUACACCUUUUUGCAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUUAAGCCGCUUGUUGU ((((((........))))))-------....-----..................(((((((.(....((((....))))....(((.......)))).))))))) ( -16.90) >DroSim_CAF1 163066 92 + 1 GUUGCCUGUUCUUUGGCAAU-------UUU------UUUUUCGUAUACCUUUUUGCAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUUAAGCCGCUUGUUGU ((((((........))))))-------...------..................(((((((.(....((((....))))....(((.......)))).))))))) ( -16.90) >DroEre_CAF1 161796 92 + 1 GUUGCCUGUUCUCUGGUAAU-------UUUUUUUCAUUUUUCGUAUACCUCUUUGCAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUCAAGCCGUU------ (((((.(((((..(((.((.-------...)).)))......(((........)))...)))))...)))))...........(((.......)))...------ ( -14.40) >DroYak_CAF1 170707 105 + 1 GUUGCCUGUUCUUUGGUACUUGGUUUUUUUUUUAUAUUUUUCGUAUAACUCUUUGCAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUUAAGCCGUUUGUUGU ..((((........)))).............((((((.....))))))......(((((((((....((((....))))....(((.......)))))))))))) ( -18.10) >DroPer_CAF1 208940 92 + 1 GCUGUGUGUA----GACGUU-GUU---GUUGUUCUUUUUGGCGGGGAAC-----GAAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUUAAACCGUUUGCUGU .......(((----((((((-(((---(((((((.(((((........)-----)))).)))))...))))))))........((.........))))))))... ( -21.50) >consensus GUUGCCUGUUCUUUGGCAAU_______UUUU_____UUUUUCGUAUACCUUUUUGCAACGAACAUUUGCAACAAUUUGUUAUUGGCCAUUUAAGCCGUUUGUUGU ((((((........))))))..................................(((((((((....((((....))))....(((.......)))))))))))) (-11.84 = -13.53 + 1.70)


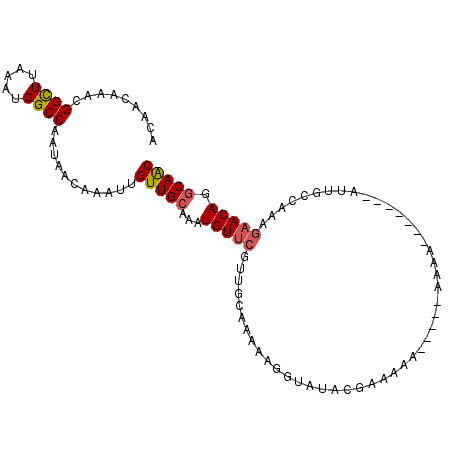
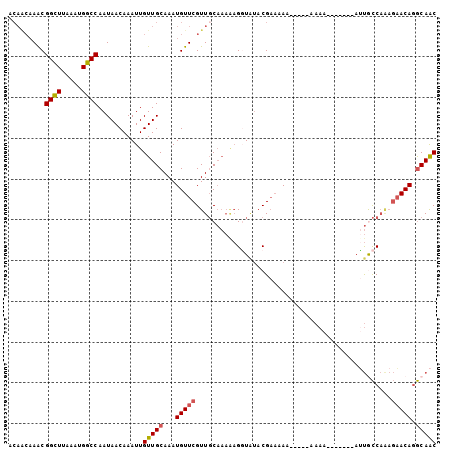
| Location | 6,419,495 – 6,419,587 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -17.12 |
| Consensus MFE | -12.37 |
| Energy contribution | -12.59 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6419495 92 - 23771897 ACAACAAACGGCUUAAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUGCAAAAAGGUAUACGAAAAA------AAA-------AUUGCCAAAGAACAGGCAAC .........((((.....))))...........(((((...(((((...((((................------...-------.))))....))))).))))) ( -18.85) >DroSec_CAF1 157504 93 - 1 ACAACAAGCGGCUUAAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUGCAAAAAGGUGUACGAAAAA-----AAAA-------AUUGCCAAAGAACAGGCAAC .........((((.....))))....(((..((.((((((.......)))))).))..)))........-----....-------.(((((........))))). ( -19.40) >DroSim_CAF1 163066 92 - 1 ACAACAAGCGGCUUAAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUGCAAAAAGGUAUACGAAAAA------AAA-------AUUGCCAAAGAACAGGCAAC .......((((((.....))))...(((.....))))).....((((((((......))).)))))...------...-------.(((((........))))). ( -19.00) >DroEre_CAF1 161796 92 - 1 ------AACGGCUUGAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUGCAAAGAGGUAUACGAAAAAUGAAAAAAA-------AUUACCAGAGAACAGGCAAC ------...((((.....))))...........(((((...(((((.((.....))((((..................-------..))))...))))).))))) ( -17.85) >DroYak_CAF1 170707 105 - 1 ACAACAAACGGCUUAAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUGCAAAGAGUUAUACGAAAAAUAUAAAAAAAAAACCAAGUACCAAAGAACAGGCAAC .........((((.....))))...........(((((...(((((.(((......(((.....................))).......))).))))).))))) ( -15.92) >DroPer_CAF1 208940 92 - 1 ACAGCAAACGGUUUAAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUUC-----GUUCCCCGCCAAAAAGAACAAC---AAC-AACGUC----UACACACAGC ...((....((((.....)))).........((((((....(((((.(((.-----((.....)).)))..))))).)---)))-))....----........)) ( -11.70) >consensus ACAACAAACGGCUUAAAUGGCCAAUAACAAAUUGUUGCAAAUGUUCGUUGCAAAAAGGUAUACGAAAAA_____AAAA_______AUUGCCAAAGAACAGGCAAC .........((((.....))))...........(((((...(((((................................................))))).))))) (-12.37 = -12.59 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:37 2006