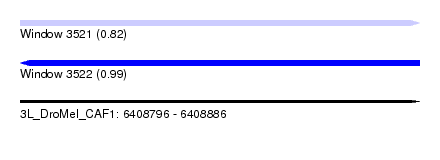
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,408,796 – 6,408,886 |
| Length | 90 |
| Max. P | 0.989393 |

| Location | 6,408,796 – 6,408,886 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -10.32 |
| Energy contribution | -12.52 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
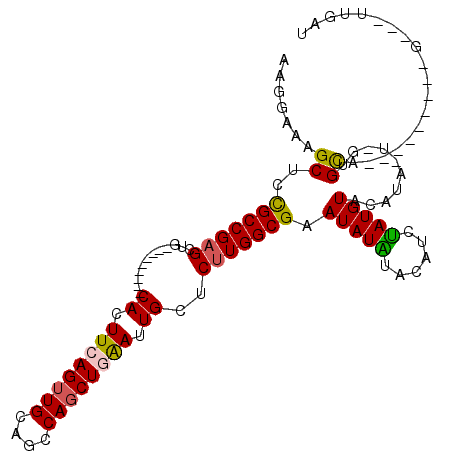
>3L_DroMel_CAF1 6408796 90 + 23771897 CACAA---C---------UGCAC-A-UAUGUACAUAGAUGUAUAUAUUCGCCAAGAGCAAUUCAGCUGGCUGCAACUGAAGUG---------CAGCUCGGCGGAGCUUUCCUU .....---.---------.((..-.-((((((((....))))))))((((((...(((......)))(((((((.(....)))---------))))).))))))))....... ( -29.00) >DroSec_CAF1 146800 90 + 1 AUCAA---C---------UGCAC-A-UAUGUACAUAGAUGUAUAUAUUCGCCAAGAGCAAUUCAGCUGGCUGCAACUGAAGUG---------CAGCUCGGCGGAGCUUUCCUU .....---.---------.((..-.-((((((((....))))))))((((((...(((......)))(((((((.(....)))---------))))).))))))))....... ( -29.00) >DroSim_CAF1 153052 90 + 1 AUCAA---C---------UGCAC-A-UAUGUACAUAGAUGUAUAUAUUCGCCAAGAGCAAUUCAGCUGGCUGCAACUGAAGUG---------CAGCUCGGCGGAGCUUUCCUU .....---.---------.((..-.-((((((((....))))))))((((((...(((......)))(((((((.(....)))---------))))).))))))))....... ( -29.00) >DroEre_CAF1 151194 89 + 1 ACCAG---C---------UACACCA-GCUACACAUAGA--UACAUAUGCGCCAAGUGCAGUCGAGCUGCCUGCAUCUGAAGUG---------CAGCUCGGCAGAGCUUUCCUU .....---.---------......(-(((.........--......(((((...)))))((((((((((((........)).)---------)))))))))..))))...... ( -29.00) >DroYak_CAF1 159758 111 + 1 GCCAUACGCCAUACUCCGUACGCCAUACUACACAUGGA--UACAUAUUCGCCAAGAGCAGUCGAGCUGCCUGCAACUGAAGUGCAGCAAGUGCAGCGCGGCAGAGCUUUCCUU ((..((((........))))..((((.......)))).--.........))..(((((.((((.((((((((((.(....)))))).....))))).))))...))))).... ( -26.50) >consensus ACCAA___C_________UGCAC_A_UAUGUACAUAGAUGUAUAUAUUCGCCAAGAGCAAUUCAGCUGGCUGCAACUGAAGUG_________CAGCUCGGCGGAGCUUUCCUU ..............................................((((((..((((..(((((.((....)).))))).((((.....))))))))))))))......... (-10.32 = -12.52 + 2.20)


| Location | 6,408,796 – 6,408,886 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.00 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6408796 90 - 23771897 AAGGAAAGCUCCGCCGAGCUG---------CACUUCAGUUGCAGCCAGCUGAAUUGCUCUUGGCGAAUAUAUACAUCUAUGUACAUA-U-GUGCA---------G---UUGUG ..(((....)))((((((..(---------((.((((((((....)))))))).))).)))))).......((((....(((((...-.-)))))---------.---.)))) ( -32.60) >DroSec_CAF1 146800 90 - 1 AAGGAAAGCUCCGCCGAGCUG---------CACUUCAGUUGCAGCCAGCUGAAUUGCUCUUGGCGAAUAUAUACAUCUAUGUACAUA-U-GUGCA---------G---UUGAU ..(((....)))((((((..(---------((.((((((((....)))))))).))).))))))..........(((..(((((...-.-)))))---------.---..))) ( -32.20) >DroSim_CAF1 153052 90 - 1 AAGGAAAGCUCCGCCGAGCUG---------CACUUCAGUUGCAGCCAGCUGAAUUGCUCUUGGCGAAUAUAUACAUCUAUGUACAUA-U-GUGCA---------G---UUGAU ..(((....)))((((((..(---------((.((((((((....)))))))).))).))))))..........(((..(((((...-.-)))))---------.---..))) ( -32.20) >DroEre_CAF1 151194 89 - 1 AAGGAAAGCUCUGCCGAGCUG---------CACUUCAGAUGCAGGCAGCUCGACUGCACUUGGCGCAUAUGUA--UCUAUGUGUAGC-UGGUGUA---------G---CUGGU ......((((..(((.(((((---------(((...(((((((.((.((.(((......)))))))...))))--)))..)))))))-))))..)---------)---))... ( -35.80) >DroYak_CAF1 159758 111 - 1 AAGGAAAGCUCUGCCGCGCUGCACUUGCUGCACUUCAGUUGCAGGCAGCUCGACUGCUCUUGGCGAAUAUGUA--UCCAUGUGUAGUAUGGCGUACGGAGUAUGGCGUAUGGC .......(((..((((..((((((.((.((((......((((.(((((.....)))))....))))...))))--..)).))))))..))))(((((........)))))))) ( -33.60) >consensus AAGGAAAGCUCCGCCGAGCUG_________CACUUCAGUUGCAGCCAGCUGAAUUGCUCUUGGCGAAUAUAUACAUCUAUGUACAUA_U_GUGCA_________G___UUGAU .......((..(((((((............((.((((((((....)))))))).))..))))))).(((((......)))))..........))................... (-16.32 = -16.00 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:31 2006