
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,367,609 – 6,367,746 |
| Length | 137 |
| Max. P | 0.986005 |

| Location | 6,367,609 – 6,367,709 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -17.34 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6367609 100 + 23771897 AGGGGCCGUACCCCAUUGAUCUCACACUAAAGCUGCUCGAUCAAGGC---UGCGAAUAGAUUUCG----AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGC .((((.....)))).(((((((((..(((..((.(((.......)))---.))...)))......----((((((......)))))).))))))))).......... ( -31.30) >DroSec_CAF1 132035 100 + 1 AGGGGCCGUACCCCAUUGAUCUCACACUAAAGCUGUUCGAUCAAGGC---UGCGAAUAGAAUUCG----AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGC .((((.....)))).(((((((((........(((((((........---..)))))))......----((((((......)))))).))))))))).......... ( -31.70) >DroSim_CAF1 138748 100 + 1 AGGGGCCGUACCCCAUUGAUCUCACACUAAAGCUGCUCGAUCAAGGC---UGCGAAUAGAUUUCG----AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGC .((((.....)))).(((((((((..(((..((.(((.......)))---.))...)))......----((((((......)))))).))))))))).......... ( -31.30) >DroEre_CAF1 136460 83 + 1 AGGGGCCGAACCCCAUUGAUCUCGUACUAAAGCUGCUC--------------------GAUUGCG----AGAUCGUUUAACCGAUCUAAGAGAUCAGCUAGAAUUGC .((((.....)))).((((((((..........(((..--------------------....)))----((((((......))))))..)))))))).......... ( -26.40) >DroYak_CAF1 144515 100 + 1 AGGGGCCGAACCCCAUUGAUCUCACACUAAAGCUGCACGUUCCAGUC---UCCGCACAGAUUUCG----AGAUCGUUUAACCGAUCUAAGAGAUCAACUAGAAUUGC .((((.....)))).((((((((..............((....((((---(......))))).))----((((((......))))))..)))))))).......... ( -26.80) >DroAna_CAF1 134381 98 + 1 GGGGGCUGGAAACCGUUGAUCUCGGCCCGGAGCUCCCCAAACAAGUCGGUUGGGUUGCGUUUUCGAAGCAGAUCGGCUGGCCGA---------UCAACAAAAAUUGC ..((........))(((((..((((((.((......)).....((((((((..(((.((....)).))).))))))))))))))---------)))))......... ( -32.30) >consensus AGGGGCCGUACCCCAUUGAUCUCACACUAAAGCUGCUCGAUCAAGGC___UGCGAAUAGAUUUCG____AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGC .((((.....)))).((((((((...................................(....).....((((((......))))))..)))))))).......... (-17.34 = -18.70 + 1.36)



| Location | 6,367,609 – 6,367,709 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -20.44 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6367609 100 - 23771897 GCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU----CGAAAUCUAUUCGCA---GCCUUGAUCGAGCAGCUUUAGUGUGAGAUCAAUGGGGUACGGCCCCU .........(((((((((((((((((......))))).----......(((...((.---(((......).)).))..)))))))))))))))((((.....)))). ( -33.10) >DroSec_CAF1 132035 100 - 1 GCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU----CGAAUUCUAUUCGCA---GCCUUGAUCGAACAGCUUUAGUGUGAGAUCAAUGGGGUACGGCCCCU .........(((((((((((((((((......)))))(----(((..((........---.....))))))..........))))))))))))((((.....)))). ( -31.52) >DroSim_CAF1 138748 100 - 1 GCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU----CGAAAUCUAUUCGCA---GCCUUGAUCGAGCAGCUUUAGUGUGAGAUCAAUGGGGUACGGCCCCU .........(((((((((((((((((......))))).----......(((...((.---(((......).)).))..)))))))))))))))((((.....)))). ( -33.10) >DroEre_CAF1 136460 83 - 1 GCAAUUCUAGCUGAUCUCUUAGAUCGGUUAAACGAUCU----CGCAAUC--------------------GAGCAGCUUUAGUACGAGAUCAAUGGGGUUCGGCCCCU .......(((((((((.....)))))))))...(((((----(((....--------------------..)).((....))..))))))...((((.....)))). ( -28.40) >DroYak_CAF1 144515 100 - 1 GCAAUUCUAGUUGAUCUCUUAGAUCGGUUAAACGAUCU----CGAAAUCUGUGCGGA---GACUGGAACGUGCAGCUUUAGUGUGAGAUCAAUGGGGUUCGGCCCCU .........(((((((((..((((((......))))))----..(((.(((..((..---........))..))).))).....)))))))))((((.....)))). ( -35.50) >DroAna_CAF1 134381 98 - 1 GCAAUUUUUGUUGA---------UCGGCCAGCCGAUCUGCUUCGAAAACGCAACCCAACCGACUUGUUUGGGGAGCUCCGGGCCGAGAUCAACGGUUUCCAGCCCCC ....((((((..((---------((((....)))))).....)))))).((..(((((.(.....).)))))..))...((((.(((((.....)))))..)))).. ( -31.70) >consensus GCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU____CGAAAUCUAUUCGCA___GCCUUGAUCGAGCAGCUUUAGUGUGAGAUCAAUGGGGUACGGCCCCU .........(((((((((..((((((......)))))).................................(((.......))))))))))))((((.....)))). (-20.44 = -21.72 + 1.28)

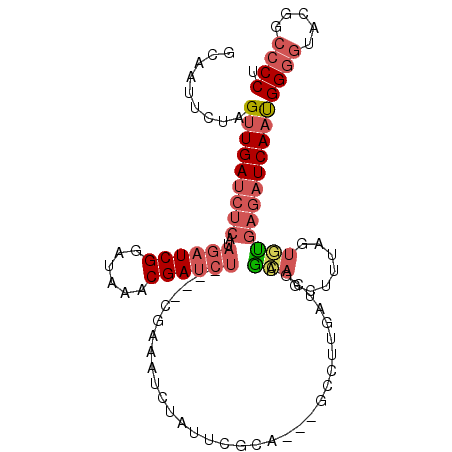
| Location | 6,367,642 – 6,367,746 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -13.09 |
| Energy contribution | -15.95 |
| Covariance contribution | 2.86 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6367642 104 + 23771897 UGCUCGAUCAAGGC---UGCGAAUAGAUUUCG----AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGCUACCAAUUCGGGGCA-GUUUAGUUAAUUAACCGAACCA ...(((....((((---(((.....(((((((----((((((......)))))).)))))))..((.((((((....)))))).)))))-)))).(((....)))))).... ( -29.70) >DroSec_CAF1 132068 104 + 1 UGUUCGAUCAAGGC---UGCGAAUAGAAUUCG----AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGCUACCAAUUCGGGGCA-GUUUAGUUAAUUAACCGAACCA .(((((....((((---(((.....((.((((----((((((......)))))).)))).))..((.((((((....)))))).)))))-)))).(((....)))))))).. ( -29.10) >DroSim_CAF1 138781 104 + 1 UGCUCGAUCAAGGC---UGCGAAUAGAUUUCG----AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGCUACCAAUUCGGGGCA-GUUUAGUUAAUUAACCGAACCA ...(((....((((---(((.....(((((((----((((((......)))))).)))))))..((.((((((....)))))).)))))-)))).(((....)))))).... ( -29.70) >DroEre_CAF1 136493 86 + 1 UGCUC--------------------GAUUGCG----AGAUCGUUUAACCGAUCUAAGAGAUCAGCUAGAAUUGCCACCGAUUCG-GGCA-GUUUAGUUAAUUAACCGAACCA (((((--------------------((((...----((((((......))))))....)))).....((((((....)))))))-))))-(((..(((....)))..))).. ( -24.80) >DroYak_CAF1 144548 104 + 1 UGCACGUUCCAGUC---UCCGCACAGAUUUCG----AGAUCGUUUAACCGAUCUAAGAGAUCAACUAGAAUUGCUACCAAUUCGUGGCA-GUUUAGUUAAUUAACCAAACCA (((((......)).---...)))..((((((.----((((((......))))))..))))))(((((((.(((((((......))))))-)))))))).............. ( -27.00) >DroAna_CAF1 134414 103 + 1 UCCCCAAACAAGUCGGUUGGGUUGCGUUUUCGAAGCAGAUCGGCUGGCCGA---------UCAACAAAAAUUGCUACCAAUUCCGCUUAGGCCUAAUUAACCAACAAAACCA ...........((..((((((((.((....))((((.((((((....))))---------))......(((((....)))))..)))).))))))))..))........... ( -25.60) >consensus UGCUCGAUCAAGGC___UGCGAAUAGAUUUCG____AGAUCGUUUAUCCGAUCUAUGAGAUCAACUAGAAUUGCUACCAAUUCGGGGCA_GUUUAGUUAAUUAACCGAACCA (((((....................((((((.....((((((......))))))..)))))).....((((((....)))))).))))).((((.(((....))).)))).. (-13.09 = -15.95 + 2.86)


| Location | 6,367,642 – 6,367,746 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -14.86 |
| Energy contribution | -16.92 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6367642 104 - 23771897 UGGUUCGGUUAAUUAACUAAAC-UGCCCCGAAUUGGUAGCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU----CGAAAUCUAUUCGCA---GCCUUGAUCGAGCA ..((((((((((.........(-(((...((((((....)))))).....(((.((..((((((......))))))----.)).))).....)))---)..)))))))))). ( -28.00) >DroSec_CAF1 132068 104 - 1 UGGUUCGGUUAAUUAACUAAAC-UGCCCCGAAUUGGUAGCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU----CGAAUUCUAUUCGCA---GCCUUGAUCGAACA ..((((((((((.........(-((((.......)))))........((((.......((((((......))))))----(((((...)))))))---)).)))))))))). ( -27.20) >DroSim_CAF1 138781 104 - 1 UGGUUCGGUUAAUUAACUAAAC-UGCCCCGAAUUGGUAGCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU----CGAAAUCUAUUCGCA---GCCUUGAUCGAGCA ..((((((((((.........(-(((...((((((....)))))).....(((.((..((((((......))))))----.)).))).....)))---)..)))))))))). ( -28.00) >DroEre_CAF1 136493 86 - 1 UGGUUCGGUUAAUUAACUAAAC-UGCC-CGAAUCGGUGGCAAUUCUAGCUGAUCUCUUAGAUCGGUUAAACGAUCU----CGCAAUC--------------------GAGCA ..((((((((............-((((-(......).))))....(((((((((.....)))))))))........----...))))--------------------)))). ( -24.50) >DroYak_CAF1 144548 104 - 1 UGGUUUGGUUAAUUAACUAAAC-UGCCACGAAUUGGUAGCAAUUCUAGUUGAUCUCUUAGAUCGGUUAAACGAUCU----CGAAAUCUGUGCGGA---GACUGGAACGUGCA .(((((((((....))))))))-)((((.....)))).(((.((((((((........((((((......))))))----.....(((....)))---))))))))..))). ( -30.60) >DroAna_CAF1 134414 103 - 1 UGGUUUUGUUGGUUAAUUAGGCCUAAGCGGAAUUGGUAGCAAUUUUUGUUGA---------UCGGCCAGCCGAUCUGCUUCGAAAACGCAACCCAACCGACUUGUUUGGGGA .(((((.(((....))).)))))...(((...((((.((((.........((---------((((....))))))))))))))...)))..(((((.(.....).))))).. ( -27.60) >consensus UGGUUCGGUUAAUUAACUAAAC_UGCCCCGAAUUGGUAGCAAUUCUAGUUGAUCUCAUAGAUCGGAUAAACGAUCU____CGAAAUCUAUUCGCA___GCCUUGAUCGAGCA ..(((((((((((((((((..........((((((....)))))))))))))......((((((......)))))).........................)))))))))). (-14.86 = -16.92 + 2.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:26 2006