
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,345,535 – 6,345,630 |
| Length | 95 |
| Max. P | 0.880675 |

| Location | 6,345,535 – 6,345,630 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 72.12 |
| Mean single sequence MFE | -16.00 |
| Consensus MFE | -8.80 |
| Energy contribution | -8.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
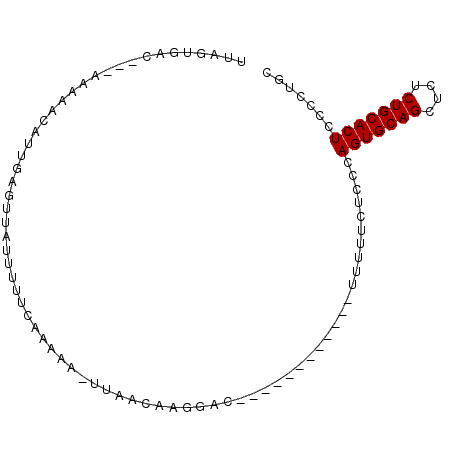
>3L_DroMel_CAF1 6345535 95 + 23771897 UUAGUAAAAAAAAAAACAUUGAGUUAUUUUUCCAAAA-UUAACAAGAACCGAAACUCUGGCUUUUUCUUCCAGUGCAGCUCUCUGCACUCCCCUGC .........(((((..((..(((((....(((.....-.......)))....)))))))..))))).....(((((((....)))))))....... ( -12.60) >DroSec_CAF1 110317 92 + 1 UUAGGGAC---AAAAACAUUGGGUUAUUUUUCAAAAU-UUCACAAUGACAAGAACUCUUGCCUUUUCUCCCAGUGCAGCUCUCUGCACUCCCCUGC ...((((.---((((.(((((.(..((((....))))-..).))))).((((....))))..)))).))))(((((((....)))))))....... ( -23.70) >DroSim_CAF1 116249 80 + 1 UUUGUGAC---AAAAACAUUAAGUUAUUUUUCUAAAA-UUAACAAGGAC------------UUUUUCUCUCAGUGCAGCUCUCUGCACUCCCCUGC ..((.((.---(((((......((((.(((....)))-.))))......------------))))).)).))((((((....))))))........ ( -11.70) >DroYak_CAF1 120784 81 + 1 GUA--UAU-CAUAACACUUAGAGUUAUGUUUCGAAAACUUCAGUUGCAG------------UUUUUCUCUCAGUGCAGCUCUCUGCACUCCCCUGC ...--...-((((((.......))))))....(((((((.(....).))------------))))).....(((((((....)))))))....... ( -16.00) >consensus UUAGUGAC___AAAAACAUUGAGUUAUUUUUCAAAAA_UUAACAAGGAC____________UUUUUCUCCCAGUGCAGCUCUCUGCACUCCCCUGC .......................................................................(((((((....)))))))....... ( -8.80 = -8.80 + -0.00)


| Location | 6,345,535 – 6,345,630 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 72.12 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -11.25 |
| Energy contribution | -10.25 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6345535 95 - 23771897 GCAGGGGAGUGCAGAGAGCUGCACUGGAAGAAAAAGCCAGAGUUUCGGUUCUUGUUAA-UUUUGGAAAAAUAACUCAAUGUUUUUUUUUUUACUAA .......(((((((....)))))))((((((((((((..(((((....((((......-....))))....)))))...))))))))))))..... ( -25.10) >DroSec_CAF1 110317 92 - 1 GCAGGGGAGUGCAGAGAGCUGCACUGGGAGAAAAGGCAAGAGUUCUUGUCAUUGUGAA-AUUUUGAAAAAUAACCCAAUGUUUUU---GUCCCUAA ...(((((((((((....))))))((((......((((((....))))))........-...((....))...))))........---.))))).. ( -28.60) >DroSim_CAF1 116249 80 - 1 GCAGGGGAGUGCAGAGAGCUGCACUGAGAGAAAAA------------GUCCUUGUUAA-UUUUAGAAAAAUAACUUAAUGUUUUU---GUCACAAA ((((((((((((((....)))))))..........------------.)))))))...-......(((((((......)))))))---........ ( -16.90) >DroYak_CAF1 120784 81 - 1 GCAGGGGAGUGCAGAGAGCUGCACUGAGAGAAAAA------------CUGCAACUGAAGUUUUCGAAACAUAACUCUAAGUGUUAUG-AUA--UAC ((((...(((((((....)))))))..........------------)))).................((((((.......))))))-...--... ( -18.92) >consensus GCAGGGGAGUGCAGAGAGCUGCACUGAGAGAAAAA____________GUCCUUGUGAA_UUUUAGAAAAAUAACUCAAUGUUUUU___GUCACUAA ...(((.(((((((....))))))).............................(((.....)))........))).................... (-11.25 = -10.25 + -1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:20 2006