
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 6,329,507 – 6,329,633 |
| Length | 126 |
| Max. P | 0.746960 |

| Location | 6,329,507 – 6,329,604 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.99 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6329507 97 + 23771897 ------GGUG----AGCUGUUGCUGAA---GCUGCUUCAGCUCCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCCCGGUCGCAGUGAUCAUCGCGUUCGC ------((((----((((((.((((((---(...)))))))...(((((......(((((.(((....))).)))))........))))))))))..)))))((....)) ( -27.84) >DroVir_CAF1 106341 92 + 1 ------GGAU----GGCUG--GCUAGA---G-AGUUAGAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCUGGUCGCAAUGAUCAUCGCGGU--C ------....----..(((--(((...---.-)))))).((((((((........(((((((((....)))))))))(((((((......)))))))..))))))))--. ( -28.80) >DroPse_CAF1 123424 104 + 1 ------GAAGCUGAAGCCGAAGCUGAAUCUGCUGCUUCAGCUGCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCCCGGUCGCAAUGAUCAUCGCGUUCUC ------..(((((((((...(((.......))))))))))))(((((........(((((.(((....))).)))))(((((((......)))))))..)))))...... ( -33.00) >DroGri_CAF1 96957 98 + 1 GGGAUGAAAU----GGCCG--GCUAGA---G-AGUUAGAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCUGGUCGCAAUGAUCAUCGCGGU--C (.(((((...----(((((--(((...---.-)))).).)))(((((((......(((((((((....)))))))))........))))))).....))))).)...--. ( -28.44) >DroAna_CAF1 94744 91 + 1 ------GAUGCUGAUGCUGAAGCUGAA---GCUGCUUCAGCUGCGAUCGUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCCCGGCCCA------AUCGCAU---- ------.........((((((((....---...))))))))((((((........(((((((((....))))))))).....((....))...------)))))).---- ( -30.30) >DroPer_CAF1 129380 104 + 1 ------GAAGCUGAAGCCGAAGCUGAAUCUGCUGCUUCAUCUGCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCCCGGUCGCAAUGAUCAUCGCGUUCUC ------(((((((((((...(((.......)))))))))..((((((((......(((((.(((....))).)))))........)))))))).........)).))).. ( -27.84) >consensus ______GAAG____AGCCGAAGCUGAA___GCUGCUUCAGCUGCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCCCGGUCGCAAUGAUCAUCGCGUU__C ...............((....((((((........))))))((((((((......(((((.(((....))).)))))........)))))))).........))...... (-16.93 = -17.99 + 1.06)

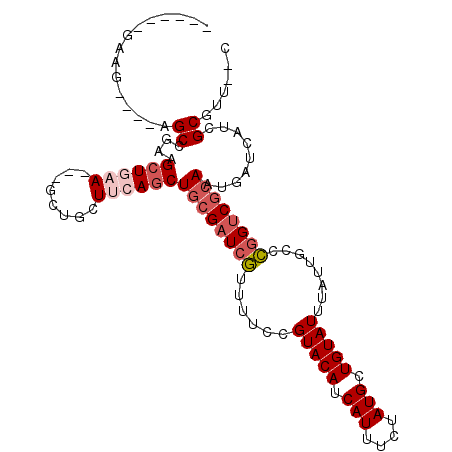

| Location | 6,329,531 – 6,329,633 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -16.00 |
| Energy contribution | -17.03 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 6329531 102 + 23771897 CAGCUCCGAUCGUUUUCCGUACAUCAUUUCUAUGCUGUAUUUAUUGCCC-GGUCGCAGUGAUCAUCGCGUUCGC------UGGGCCUCCAUUUCCGUUUCUGCGACUGC ...........((.....(((((.(((....))).))))).....)).(-((((((((.(((....((....))------(((....))).....))).))))))))). ( -22.90) >DroVir_CAF1 106362 98 + 1 GAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCU-GGUCGCAAUGAUCAUCGCGGU--C------UGGGCCUCCAUUUCCGUUGCUC--ACUGC ((((((((((((......(((((((((....)))))))))........)-))))))).........((((.--.------(((....)))...)))).))))--..... ( -31.74) >DroGri_CAF1 96984 98 + 1 GAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCU-GGUCGCAAUGAUCAUCGCGGU--C------UGGGCCUCCAUUUCCGUUGUUC--ACCUC ((((((((((((......(((((((((....)))))))))........)-))))))).........((((.--.------(((....)))...)))).))))--..... ( -29.34) >DroWil_CAF1 114771 109 + 1 GAGCUACGAUAAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCCUGGUCGCAAUGAUCAUCGCAUUGCCAGCUUUGGGGCCUCCAUUUCCGUUUUCUUGACUGC (((..(((..........(((((((((....))))))))).....((((((((.((((((.......))))))..)))..))))).........))).)))........ ( -26.70) >DroMoj_CAF1 104441 98 + 1 GAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCU-GGUCGCAAUGAUCAUCGCGCU--C------UGUGCCUCCAUUUCCGCUGUAC--AGUGU ((((.(((((........(((((((((....)))))))))(((((((..-....)))))))..))))))))--)------..............((((....--)))). ( -28.60) >DroAna_CAF1 94772 92 + 1 CAGCUGCGAUCGUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCC-CGGCCCA------AUCGCAU----------UGGGCCUCCAUUUCCGUUUCUGCGGCUGC ((((((((...((.....(((((((((....))))))))).....))..-.((((((------((...))----------))))))..............)))))))). ( -33.50) >consensus GAGCUGCGAUCAUUUUCCGUACAUCAUUUCUAUGGUGUAUUUAUUGCCC_GGUCGCAAUGAUCAUCGCGUU__C______UGGGCCUCCAUUUCCGUUGCUC__ACUGC .(((((((((........(((((((((....)))))))))(((((((.......)))))))..))))))............(((........))))))........... (-16.00 = -17.03 + 1.03)

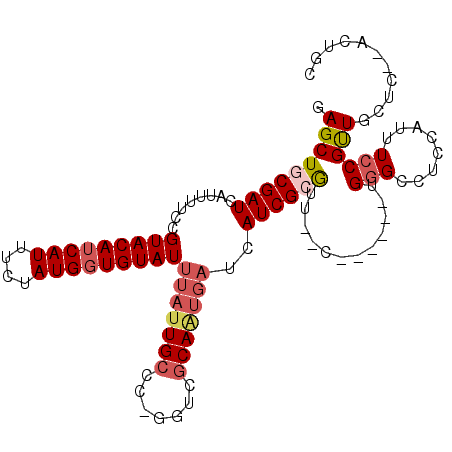
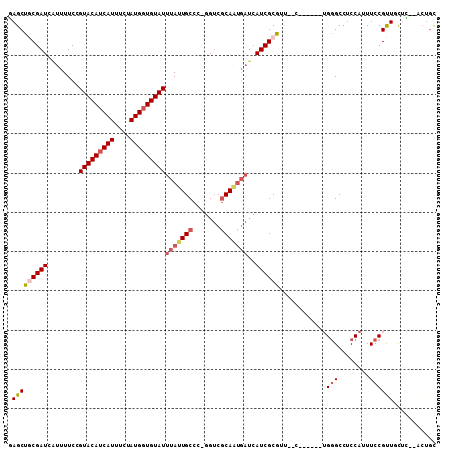
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:14 2006